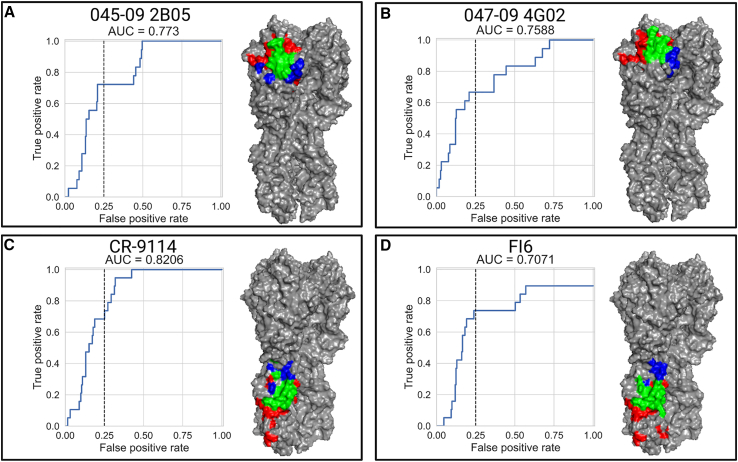
Figure 5.
Inferring mAb epitope footprints using the mAb-Patch algorithm
We used 4 mAbs with known epitopes based on solved structures of the antibodies in complex with the influenza H1 protein (047-09 4G02, 045-09 2B05, CR-9114, and FI6). The binding profiles were used to define a position-specific score, which estimates the likelihood of the position to belong to the antibody binding epitope (see STAR Methods and Table S2 for strains used in MSA). The score was used to rank all sites on the HA stalk or head. Epitope patch predictions were based on the ranked positions and their proximal neighborhoods in 3D space (see also Figure S5). For each mAb, we calculated the receiver operating characteristic (ROC) curves for epitope position ranked by mAb-Patch. The dashed line represents 25% false positive (FP) rate. The predicted vs. experimentally measured binding footprints of each mAb using a 25% FP rate are visualized on the HA 3D structure of pH1N1 (PDB: 7MEM). Positions colored in green represent positions that were correctly identified (true positives [TPs]), positions in blue represent positions that are part of the solved footprint that were not predicted by our method (false negatives [FNs]), and positions in red represent positions that are not part of the known binding footprint but were identified by our score (FPs).