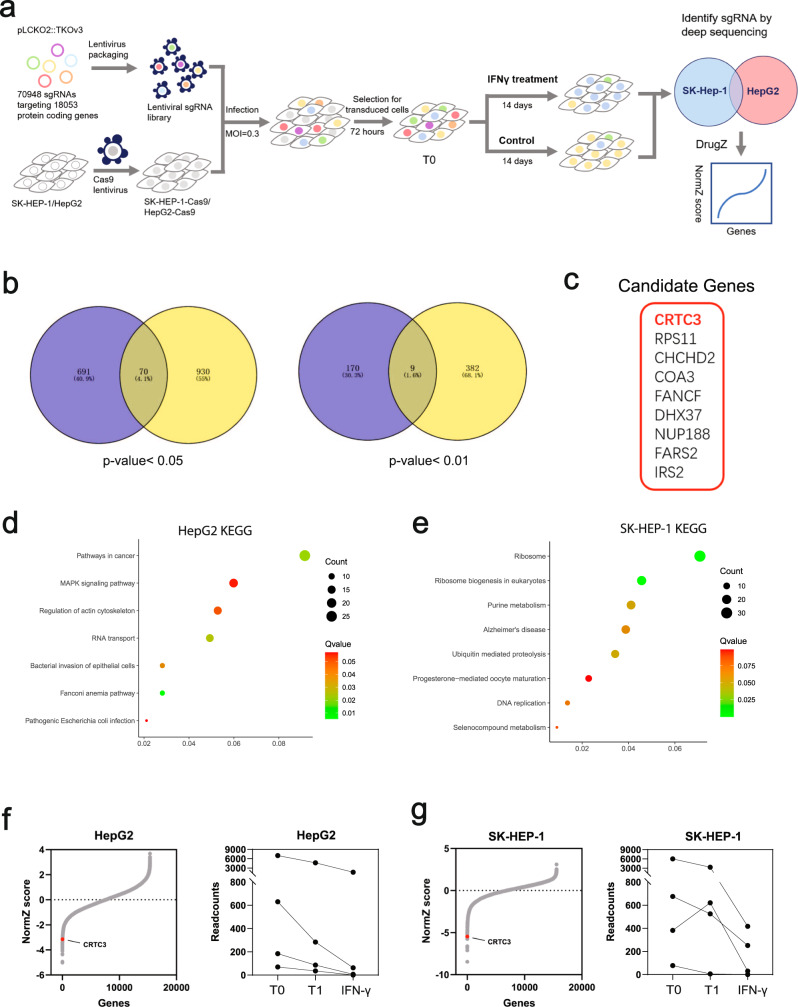
Fig. 1. CRISPR/Cas9 screening identified driver genes that were associated with IFN-γ resistance in HCC.
a General workflow of the CRISPR/Cas9 knockout library screening in this study. b Venn diagrams displaying CRISPR screening results. There were 70 significantly expressed resistance inducers, defined as P < 0.05 and 9 top resistance inducers, defined as P < 0.01 when intersecting HepG2 and SK-Hep-1 cell lines. c Nine top resisters identified as candidate genes of the screening. d, e KEGG enrichment analyses for significant gene hits in HepG2 cells and SK-HEP-1 cells. f, g The synergistic or suppressing effects on drug treatment were determined using the DrugZ algorithm and NormZ scores. The abundance of sgRNAs as the readout of library screening was analyzed using Bowtie2. CRTC3 was a significant depleted gene hit in both cell lines. The amount of sgRNA targeting CRTC3 was markedly decreased in CRTC3-KO HCC cells when treated with IFN-γ, compared to the control (T1).