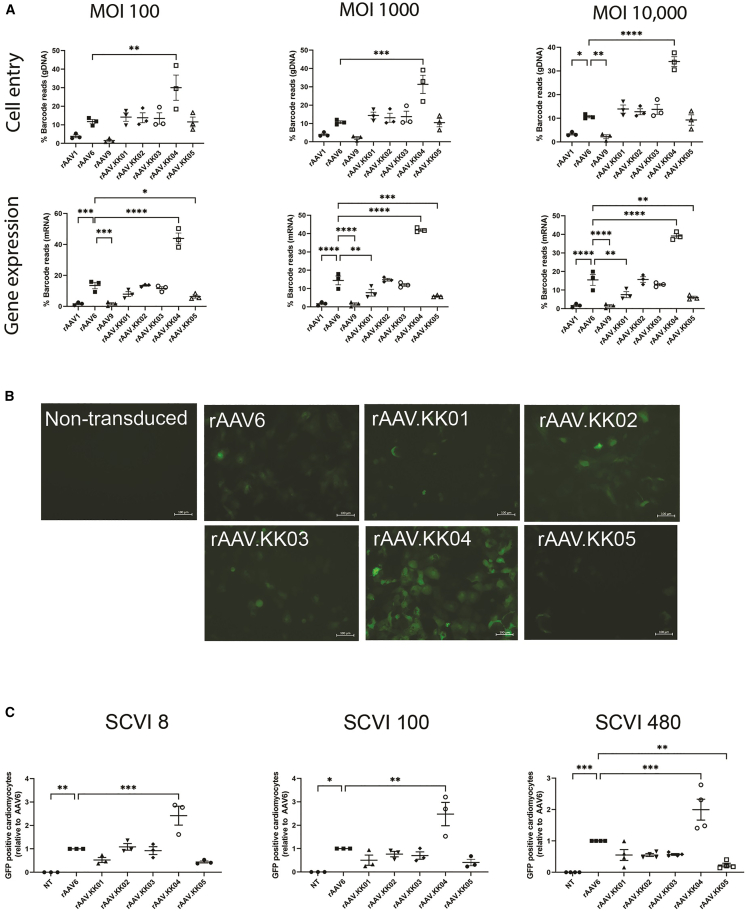
Figure 2.
rAAV.KK04 was most efficient at gene delivery to hiPSC-CMs
(A) hiPSC-CMs competitively transduced with a barcoded library of rAAV.CMV.GFP vectors, then harvested at D5 post transduction. Extraction of DNA/RNA was performed, followed by analysis using next-generation sequencing (n = 3). The relative proportions of barcode reads post NGS analysis are given at the level of cell entry (gDNA) and gene expression (mRNA) for MOT 100, 1,000, and 10,000. Results are expressed as percentage of total reads for each cell line. (B) hiPSC-CMs were transduced with unbarcoded rAAV.CBA.GFP vectors at MOT 1,000, followed by analysis using microscopy and flow cytometry to quantify GFP (n = 3 per group for SCVI 8 and 100, n = 4 per group for SCVI 480) on day 5 post transduction. Fluorescence images showing GFP auto-fluorescence in live cells. Scale bars, 100 μM. (C) Flow cytometry dot plots quantifying proportions of GFP-positive cardiomyocytes (cTnT+ cells) in three hiPSC lines. Data are mean ± SEM. Statistical significance of differences was calculated using an ordinary one-way ANOVA, and the difference between the mean of the variants and AAV6 was calculated with Dunnett’s multiple comparison test (∗ p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001, ∗∗∗∗ p ≤ 0.0001).