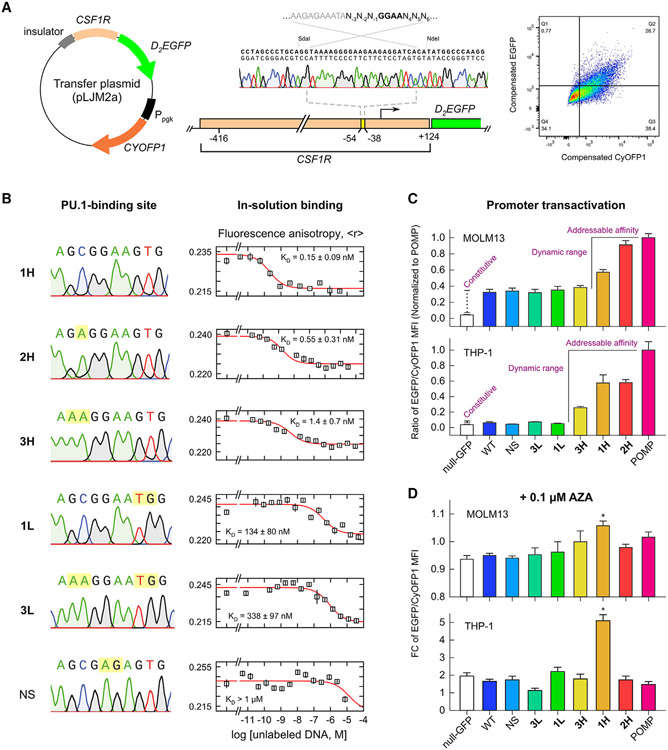
Figure 1. Affinity-dependent transactivation of the CSF1R promoter by PU.1.
(A) Design of a synthetic fluorescent CSF1R reporter. The essential PU.1-binding site was replaced by a probe-binding site. Promoter transactivation leads to expression of D2EGFP. A constitutive CyOFP1 marker affords gating of transduced cells and normalization.
(B) Panel of PU.1-binding sites in order from highest to lowest affinity. Points represent mean ± SD of three technical replicates.
(C) Promoter transactivation in MOLM-13 and THP-1 cells as mean ratios of D2EGFP/CyOFP1 fluorescence ±SE of at least three biological replicates. Signal dispersion was significant by one-way ANOVA (p < 10−6). Bracketed sequences generate significantly higher fluorescence than the constitutive intensity (p < 0.05, post hoc Tukey honestly significant difference [HSD]).
(D) Response of promoter signal to the hypomethylating agent 5-azacytidine (AZA). Shown is mean fold change in EGFP/CyOFP1 intensity ±SE of at least three biological replicates. Asterisk (*) indicates significantly above the null-GFP control (p < 0.05).