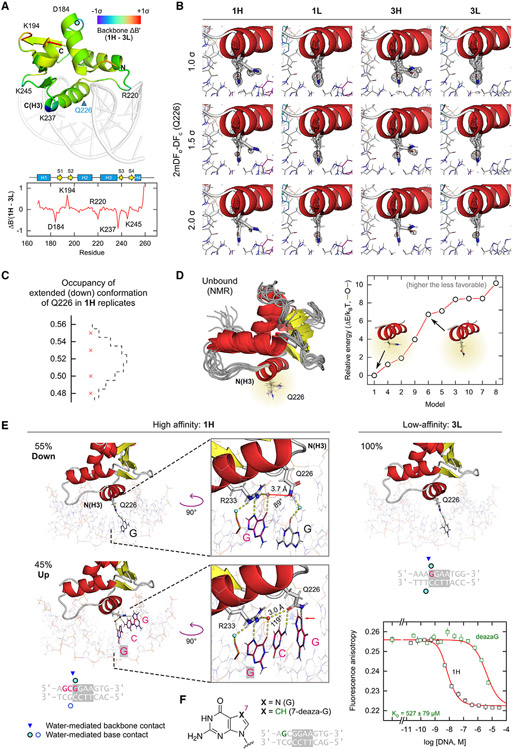
Figure 3. Alternate conformations of a critical glutamine residue in the recognition helix marks high-affinity DNA binding by PU.1.
(A) Comparison of the backbone B factors for ΔN165 in the high- and low-affinity complexes 1H and 3L. B factors are normalized to Z scores (B' factors) and their differences mapped to the structure. Internal residues with the most divergent B' factors are labeled. See also Figure S3.
(B) Sidechain conformations of Q226 with 2mFo-DFc maps at the indicated cutoffs. In addition to excess disconnected electron densities, the density around Q226 in high-affinity complexes diminishes more markedly with increasing σ than low-affinity counterparts. See also Figure S4.
(C) Occupancy of the down conformation of the Q226 sidechain in four independent co-crystals of 1H. The dashed envelope is a binomial fit to the data.
(D) The solution NMR ensemble of the unbound ETS domain (PDB: 5W3G), consisting of 10 models ranging in conformation for Q226. The relative conformational energies of the models were estimated by molecular mechanics methods and sorted by energy.
(E) Interactions of alternate Q226 conformations with DNA. In 1H, the up conformation connects both core and flanking bases in a network involving R233, but the down position does not present compatible geometry to interact with R233.
(F) Replacement of G at position −2 with 7-deaza-G in the 1H sequence, which denies H-bonding at position 7, reduced binding to low-affinity levels. Points represent mean ± SD of three technical replicates.