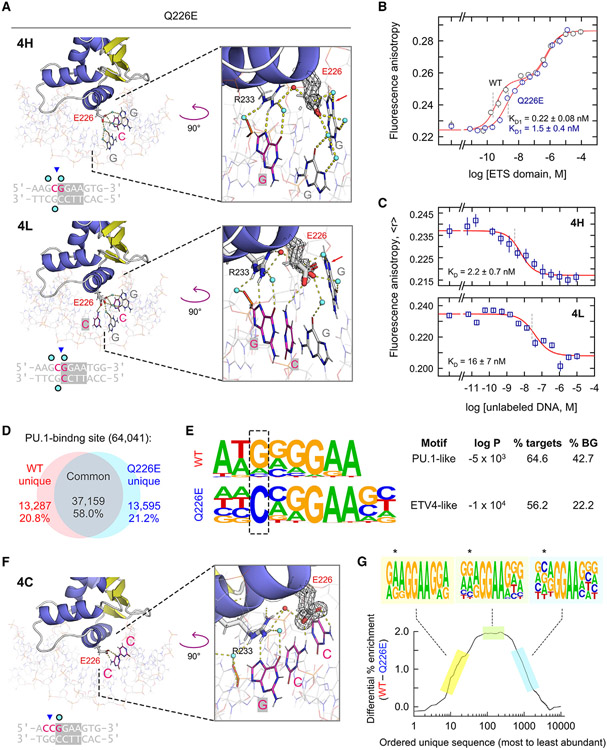
Figure 4. The Q226E mutation fundamentally alters DNA selection by PU.1.
(A) ΔN165(Q226E) in complex with the high- and low-affinity DNA 1H and 1L, respectively. In 4L, E226 exhibited occupancies best fitted by down conformations. 2mFo-DFc maps are rendered at 1.0 σ. Arrows mark the N7 atom of G−2. See also Figures S5A, S5C, and S6.
(B) Direct DNA binding by WT ΔN165 and Q226E. Points represent mean ± SD of three technical replicates.
(C) Competition titrations comparing Q226E binding to high- and low-affinity DNA. Points represent mean ± SD of three technical replicates.
(D) Summary of genomic localization of full-length WT and Q226E PU.1 in HEK293T cells.
(E) The most highly enriched motifs bound by WT and Q226E from a de novo motif analysis. The −2 position flanking the 5' end of the core consensus is boxed. Affordance for cytosine at this position is characteristic of other ETS members such as the class II ETV subfamily.6
(F) Q226E in complex with 5'-flanking cytosines. The E226 sidechain shows full occupancy in the up conformation and contacts the exocyclic NH2 of cytosines. See also Figure S5D.
(G) Enrichment of sites containing 5'-GGAA-3' in WT-bound relative to Q226E-bound genomes. Unique sequences were sorted in decreasing order of abundance. The ordinate represents the difference of the ordered sequence counts for WT PU.1 over Q226E. The motifs summarize the subset of WT sequences indicated by the highlighted colors. The −2 position is marked with asterisks.