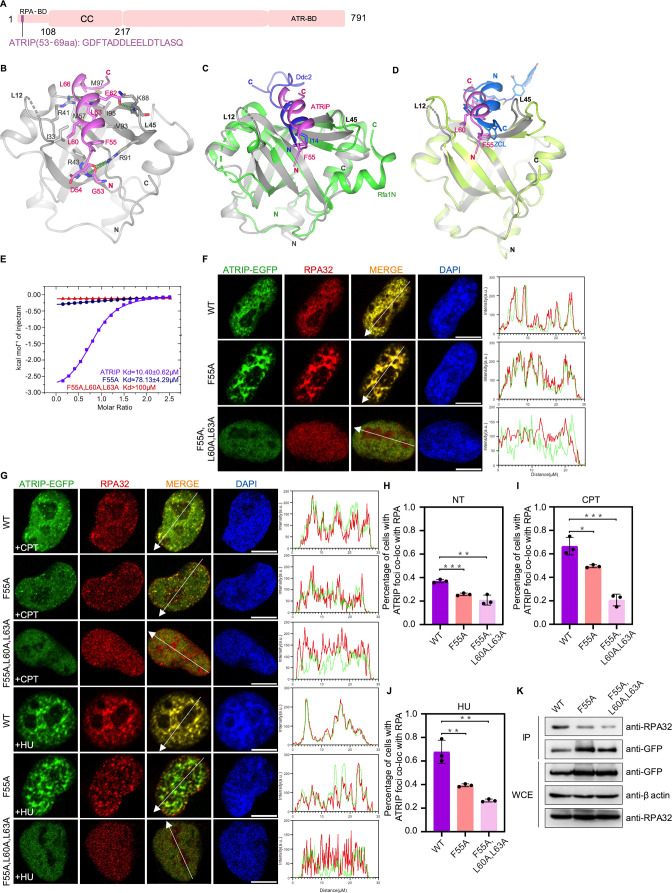
Figure 6. Structure of the RPA70N–ATRIP complex.
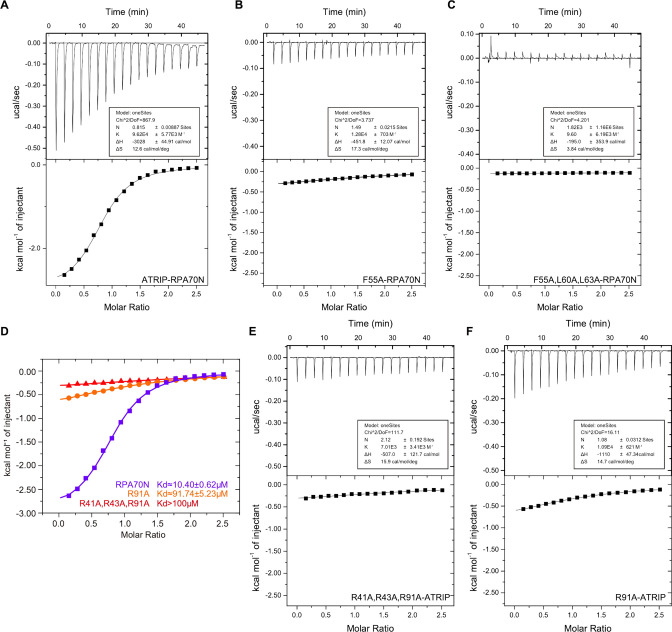
(A) Linear domain diagram of ATRIP, showing the position and sequence of the RPA70N-interacting motif. CC, coiled-coiled domain; ATR-BD, ATR binding domain. (B) Ribbon representation of the RPA70N–ATRIP crystal structure. The ATRIP peptide is colored in violet and RPA70N in light grey. Important interacting residues are shown as sticks, and green dashed lines indicate hydrogen bonds or salt bridges. (C) Alignment of the RPA70N–ATRIP structure with the Ddc2–Rfa1N structure (PDB: 5OMB), showing that ATRIP and Ddc2 bind to RPA70N in the same direction. RPA70N–ATRIP components are colored as in (B), Ddc2 is colored in green and Rfa1N is colored in blue. (D) Superposition of the RPA70N–ATRIP structure with that of the RPA70N-stapled peptide complex (PDB:4NB3). For 4NB3, RPA70N is colored in light-green and the stapled peptide is colored in marine. ZCL is 3,4-dichloro-substituted phenylalanine. The direction of the stapled peptide is reversed when compared to that of ATRIP. (E) ITC titration data for WT ATRIP (53–69 aa) and for F55A or F55A/L60A/L63A mutant peptides with RPA70N. (F, G) HeLa cells expressing ATRIP–EGFP, ATRIP (F55A)–EGFP, or ATRIP (F55A/L60A/L63A)–EGFP were treated with medium control (NT), CPT (2 μM, 2 h) or HU (2 mM, 3 h), fixed and immunostained with an anti-RPA32 antibody. The scale bar is 10 μm. The intensities of the fluorescent signals for ATRIP–EGFP, ATRIP (F55A)–EGFP or ATRIP (F55A/L60A/L63A)–EGFP and RPA32 are displayed on the right. (H, I and J) Quantification of data from (F) and (G), data are presented as mean ± s.d. of three independent experiments. 100 cells from each experiment were analyzed, and cells containing more than three bright ATRIP and RPA co-localization foci were defined as positive. Statistical analysis was performed using a two-tailed Student’s t-test (**** P<0.0001, *** P<0.001, ** P<0.01, * P<0.05). (K) Immunoprecipitation and western blots showed that mutation of ATRIP residues reduced RPA association. Anti-EGFP magnetic beads were used to carry out immunoprecipitations, which were followed by probing with an anti-RPA32 antibody. IP, immunoprecipitation; WCE, whole cell extract.