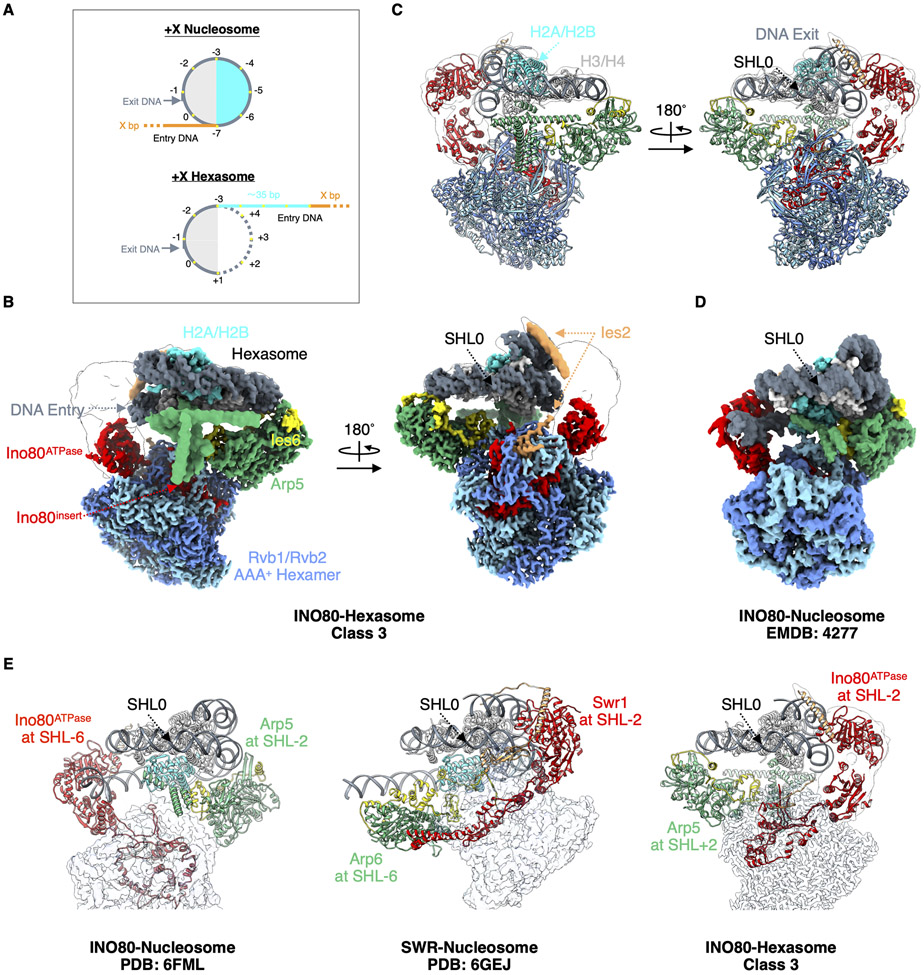
Fig. 1. Structure of the INO80-hexasome complex reveals large rotation.
(A) Cartoon illustration of a +X Nucleosome and a +X Hexasome. H2A-H2B dimer proximal to the flanking DNA (entry side dimer): cyan; H3-H4: light gray; 601 DNA: dark gray; flanking DNA: orange; additional free (unwrapped) DNA: cyan; super helical locations: yellow dots; DNA from the bottom gyre: dotted line. (B) Two different views of cryo-EM density map of the INO80-hexasome complex (class 3). (C) Atomic model of the INO80-hexasome complex (class 3), viewed in the same orientation as the map is viewed in (B). (D) Cryo-EM density map of Chaetomium thermophilum INO80-nucleosome complex (EMDB: 4277 (14)) displayed with its nucleosome dyad and H3-H4 tetramer aligned with that of the hexasome in the right panel of (B). Note that INO80 on a hexasome rotates ~180° from where it sits on a nucleosome when keeping the nucleosome/hexasome dyad and H3-H4 aligned. (E) Structural comparisons of INO80-nucleosome complex (left), SWR-nucleosome complex (middle) and INO80-hexasome complex (right), with nucleosome/hexasome dyad and H3-H4 aligned.