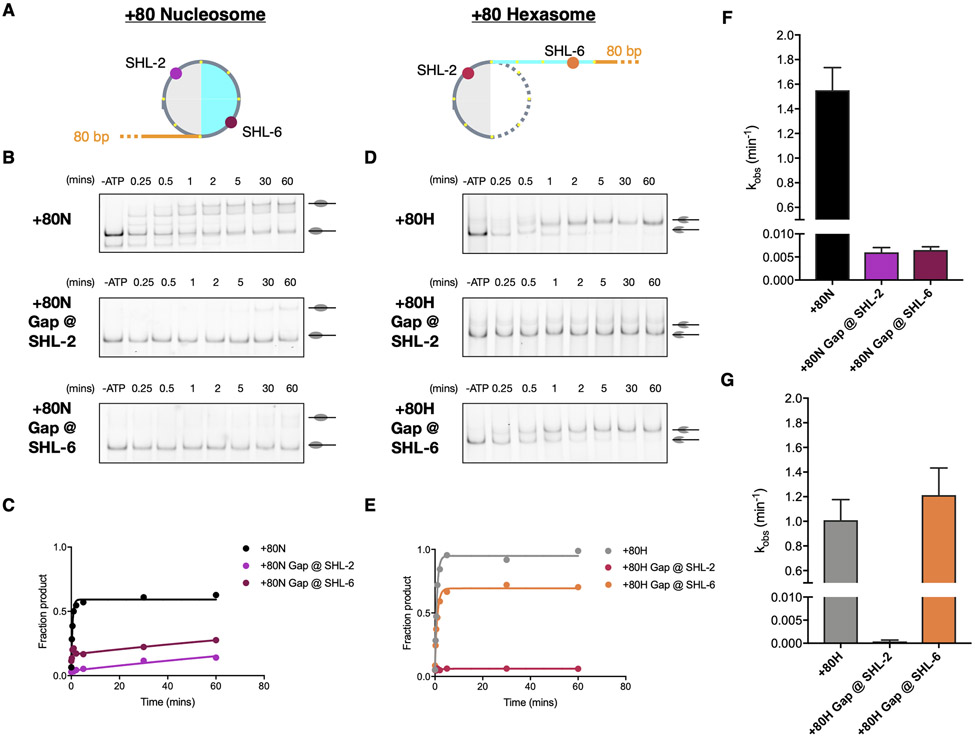
Fig. 3. Inhibition of DNA translocation at specific SHL sites influence nucleosome and hexasome sliding by INO80.
(A) Cartoon illustration of a +80 Nucleosome (left) and a +80 Hexasome (right) with approximate locations of site-specific single base gaps indicated. Colors are the same as in Fig. 1A. (B-C) Example gels and time courses of native gel-based remodeling assays of WT INO80 on +80 nucleosomes with no gap, gap near SHL-2, and gap near SHL-6. (D-E) Example gels and time courses of native gel-based remodeling assays of WT INO80 on +80 hexasomes with no gap, gap near SHL-2, and gap near SHL-6. (F-G) Average observed rate constants of INO80 sliding activity. kobs (min−1): +80N: 1.551 ± 0.1846; +80N Gap @ SHL-2: 0.005995 ± 0.001054; +80N Gap @ SHL-6: 0.006497 ± 0.0007117; +80H: 1.01 ± 0.1668; +80H Gap @ SHL-2: 0.000379 ± 0.0002849; +80H Gap @ SHL-6: 1.213 ± 0.2209. Data represent the mean ± SEM for three technical replicates performed under single-turnover conditions with saturating enzyme and ATP.