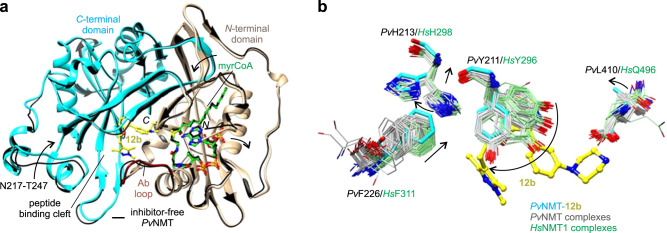
Fig. 2. Unique inhibitor binding site architecture exhibited by the PvNMT–12b complex.
a Overall architecture. 12b binds in the substrate peptide binding cleft at the interface of the NMT N- and C-terminal domains (tan, residues 1–204, truncated at residue 27, and cyan, residues 205–410, respectively), with the Ab loop (dark red, residues 95–102) in the closed conformation. Comparison with the inhibitor-free state [thin black ribbons, PDB entry 4B1019] shows significant conformational changes in the substrate binding sites (indicated schematically with arrows). b Distinct conformational changes in selected residues, namely PvTyr211, PvHis213, PvPhe226, and PvLeu410. Carbon atoms of the following crystal structures of PvNMT and HsNMT1, which were superimposed onto the structure of PvNMT-12b using Chimera51, are colored gray and light green, respectively: 2YNC/D/E20, 3IU1, 3IU2, 3IWE, 3JTK, 4A9523, 4B10/1/2/3/419, 4BBH53, 4C2Y/Z54, 4C6855, 4CAE/F22, 4UFV/W/X56, 5G1Z/2257, 5MU6/O48/O4V/O6H/O6J31, 5NPQ, 5O9S/T/U/V58, 5UUT59, 5V0W, 5V0X, 6B1L, 6FZ2/3/560, 6MB121, 6NXG18, 6PAV61, 6EHJ/QRM/SJZ/SK262, 6TW5, 6TW6, and 7RK3.