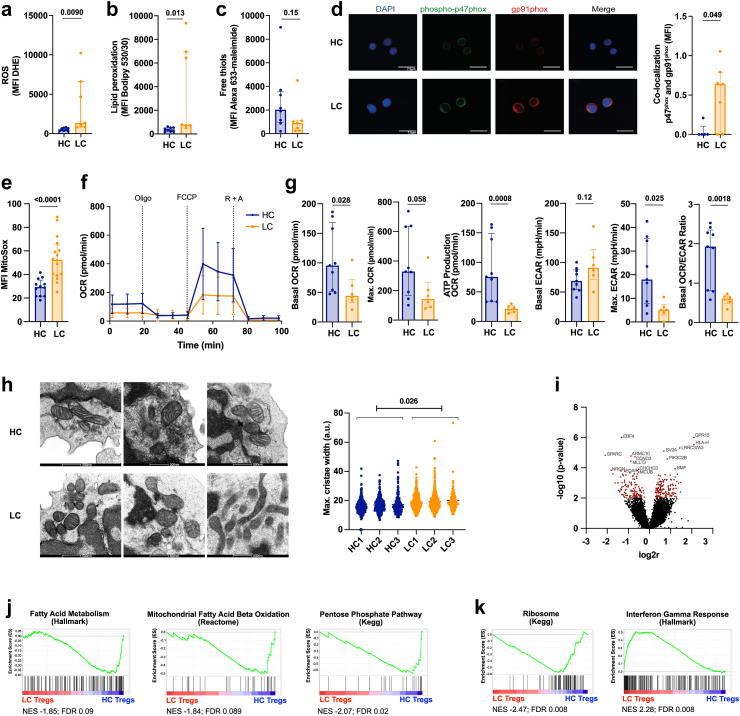
Fig. 2.
Circulating Tregs from LC patients exhibit increased intracellular ROS and changes in mitochondria morphology. a–c. Measurement of oxidative stress in freshly isolated HC and LC CD4+CD25+ Tregs. HC n = 8, LC n = 7; unpaired t-test with Welch's correction (a,b) following log10 transformation (a–c). Median (IQR). a. ROS shown by the MFI of dihydroethidium (DHE). b. Analysis of lipid peroxidation using the MFI of Bodipy C11 581/591. c. Free thiols on the cell surface indicated by the MFI of Alexa 633-maleimide. d. Representative confocal microscopy pictures (left) and summary (right) indicating co-localisation of the NADPH oxidase 2 (Nox2) subunits p47phox (green) and gp91phox (red) measured as MFI of co-localised points (white). Cell nuclei stained with DAPI (blue). Scale bar 7.5 μm. HC n = 5, LC n = 7. Mann–Whitney test. Median (IQR). e. Mitochondrial ROS measured by Mitosox (MFI) in HC and LC Tregs. HC n = 12, LC n = 16. Unpaired t-test following log10 transformation. Median (IQR). f. Metabolic profile of CD4+ CD25+ Tregs of HC and LC patients. Oxygen Consumption Rate (OCR) and extracellular acidification rate (ECAR) was measured following the exposure to Oligomycin (Oligo), FCCP and Rotenone + Antimycin A (R + A). SD. g. Basal and maximal OCR and ATP production (measured after Oligomycin treatment) in HC and LC Tregs. Basal ECAR, glycolytic capacity (max. ECAR) and basal OCR/ECAR ratio in HC and LC Tregs. HC n = 9, LC n = 6. Unpaired t-tests following log10 transformation. Median (IQR). h. Quantification of mitochondrial cristae width of HC and LC Tregs by electron microscopy. Scale bar 500 nm. HC/LC n = 3. Mean mitochondrial cristae width, HC; 16.65 (95% Confidence Interval 15.48–17.81) vs LC; 19.27 (95% Confidence Interval 18.08–20.45; p = 0.013), from linear mixed effect model (dependent variable, cristae width; fixed-effects variable, disease state; random effects variable, patient ID. Sensitivity analysis with log10 transformed cristae width p = 0.026). i. Volcano plot indicating the 15 top differentially expressed genes in LC Tregs. Genes with a p-value <0.01 were considered differentially expressed between LC and HC Tregs and marked in red. n = 7. j. Gene set enrichment analysis (GSEA) of fatty acid metabolism, fatty acid beta oxidation, and Pentose Phosphate Pathway underrepresented in LC Tregs. n = 7. k. GSEA of Ribosome and Interferon Gamma Response pathway genes in LC Tregs. n = 7. Abbreviations: Reactive oxygen species (ROS), Healthy control (HC), liver cirrhosis (LC), dihydroethidium (DHE), 4.6-diamino-2-phenylin-dole (DAPI), NADPH oxidase 2 (Nox2), Oxygen consumption rate (OCR), Extracellular acidification rate (ECAR), Oligomycin (Oligo), Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), Rotenone + Antimycin A (R + A).