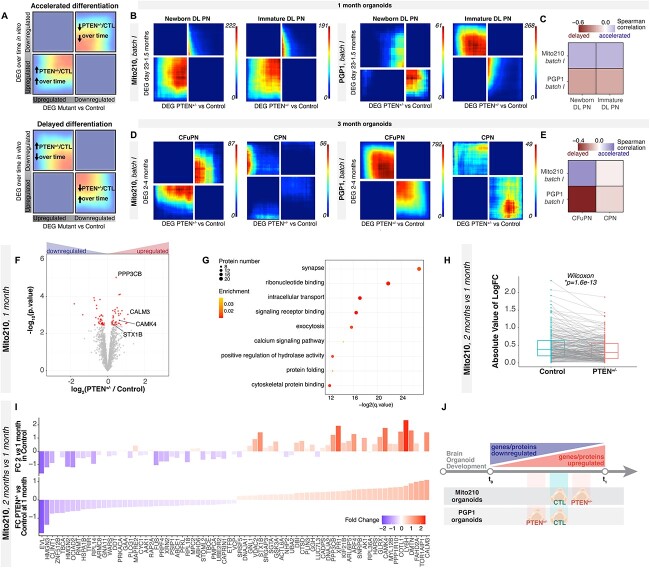
Figure 2.
Deep-layer neurons show asynchronous development across PTEN mutant organoids at multiple timepoints. (A) Schematic explaining the rank–rank hypergeometric overlap (RRHO2) output plot. Genes from each list are ordered from most upregulated to most downregulated, with the most upregulated gene of each differential gene list in the lower left corner. (B) RRHO2 plots comparing differentially expressed genes (DEG) between control and PTEN heterozygous mutant organoids at 1 month in vitro versus genes changing over time in organoids between 23 days and 1.5 months in vitro, in the newborn DLPN and immature DLPN populations. (C) Spearman correlation of DEG between each cortical cell type within control and PTEN mutant organoids at 1 month, and genes that change within that cell type from 23 days to 1.5 months in control organoids (17). Correlation is calculated on each gene’s signed logFC. (D) RRHO2 plots comparing differentially expressed genes between control and PTEN heterozygous mutant organoids at three month in vitro versus genes changing over time in organoids between two and four months in vitro, in the CFuPN and CPN populations. (E) Spearman correlation of DEG between each cortical cell type within control and PTEN heterozygous mutant organoids at 3 months, and genes that change within that cell type from 2 to 4 months in control organoids (17). Correlation is calculated on each gene’s signed logFC. (F) Volcano plot showing fold change versus FDR of measured proteins in MS experiments comparing PTEN heterozygous versus control organoids cultured for 35 days (n = 4 single organoids per genotype). Significant DEPs are shown in red (FDR < 0.1). Proteins mentioned in the text are highlighted. (G) Enriched GO terms for DEPs between PTEN heterozygous and control organoids cultured for 1 month in vitro. (H) Protein expression changes at 2 vs. 1 month for control and mutant organoids. Gray lines connect values for the same protein in the two genotypes. P-value from a paired signed Wilcoxon rank test. Only significant DEPs are shown; for the same analysis done with all detected proteins, see Supplementary Material, Figure 3. (I) Comparison of protein expression changes in PTEN heterozygous vs. control organoids at 1 month (bottom) versus changes in 2- vs. 1-month control organoids (top). Color and y-axis indicate log2 fold change. Only proteins with significantly differential expression between control and mutant at 1 month are shown (n = 3 single organoids per genotype for 2 months). (J) Schematic summarizing line-specific developmental acceleration/deceleration phenotypes from scRNA-seq and proteomics experiments. DL, deep layer; CPN, callosal projection neurons; CFuPN, corticofugal; CTL, control; FC, fold change.