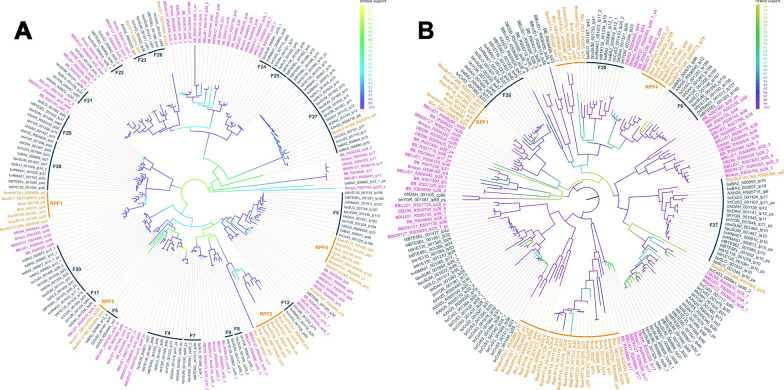
Fig. 5.
Maximum likelihood phylogenies of REP Borrelia plasmids based on PF32 (A) and PF57/62 (B) nucleotide sequences among a subset of Lyme borreliosis (LB) and relapsing fever (RF) sequences as used by Jones et al. [31] to define RF plasmid compatibility groups. Branches are colour coded according to bootstrap support and taxa labels are colour coded according to their phylogenetic lineage [cyan (LB), purple (RF), orange (REP)]. The outer rings in A show the plasmid families based on analysis of PF32 sequences, with purple rings indicating RF plasmid families (F4-29) as defined by Jones et al. [31] and orange rings indicating REP plasmid families (RPF1-4) as defined by our analysis. Labelled outer rings in B show plasmid families that are concordant between PF32 and PF57/72 loci, and unlabelled outer rings indicate putative REP plasmid families that were disconcordant with PF32 loci or for which no PF32 loci existed. The isolate prefixes, gene locus, and plasmid are given for each PF32 and PF57/62 locus. See Supplementary Information for details of isolate prefixes