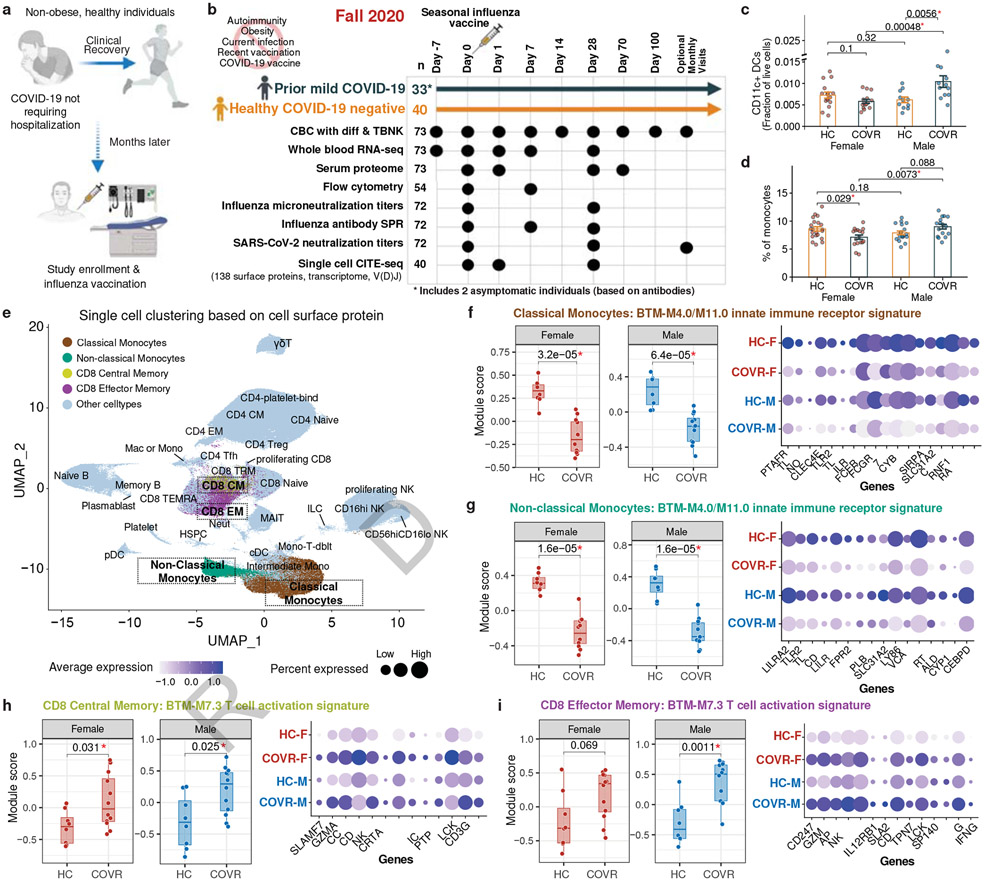
Figure 1. Study overview and baseline differences.
a, Schematic showing the study concept and design.
b, Data generated in the study. Both COVID-19-recovered (COVR) subjects and healthy controls (HC) were enrolled at seven days before vaccination (Day −7) and sampled at the indicated timepoints relative to the day of influenza vaccination. The number of subjects assayed for each data type is indicated. CBC with diff = complete blood count with differential; TBNK = T- and B-lymphocyte and Natural Killer cell phenotyping; SPR = Surface plasmon resonance.
c, Bar plots comparing the proportion of CD11c+ dendritic cells (DCs; as the fraction of live cells from flow cytometry) between COVR females (COVR-F; n=15), HC females (HC-F; n=16), COVR males (COVR-M; n=12), and HC males (HC-M; n=11) at day 0 (D0). The statistical significance is determined by two-tailed Wilcoxon test. Error bars indicate the standard error of each group.
d, Similar to (c) but for monocytes (from CBC; y-axis) between COVR-F (n=17), COVR-M (n=16), HC-F (n=21), and HC-M (n=19) at baseline (average of Day −7 and D0).
e, UMAP of the CITE-seq single cell data showing clustering of cells based on the expression of cell surface protein markers (632,100 single cells from all timepoints with CITE-seq data: days 0, 1, 28). Colored and boxed cell clusters are further explored in (f-i).
f, (left) Box plots comparing the innate immune receptor signature scores (see Methods) between HC-F (n=8) and COVR-F (n=12) (left box) and HC-M (n=8) and COVR-M (n=12) (right box) using the CITE-seq classical monocyte pseudobulk expression data at D0. Each point represents a subject. (right) Bubble plot showing the average gene expression of selected genes, including those in the Gene Ontology (GO) “pattern recognition receptor activity” and “immune receptor activity” gene sets.
g, Similar to (f) but showing the non-classical monocyte population at D0.
h, Similar to (f) but showing the T-cell activation (BTM-M7.3) module scores of CD8+ central memory T cells at D0. Bubble plot showing the average gene expression of the selected genes shared by male and female from the gene set enrichment analysis (see Methods).
i, Similar to (h) but showing the CD8+ T-cell effector memory population at D0.
All box plots show the median, first and third quantiles (lower and upper hinges) and smallest [lower hinge – 1.5× interquartile range (IQR)] and largest (upper hinge + 1.5× IQR) values (lower and upper whiskers). Unless otherwise noted, statistical significance of difference between groups is determined by two-tailed Wilcoxon test.