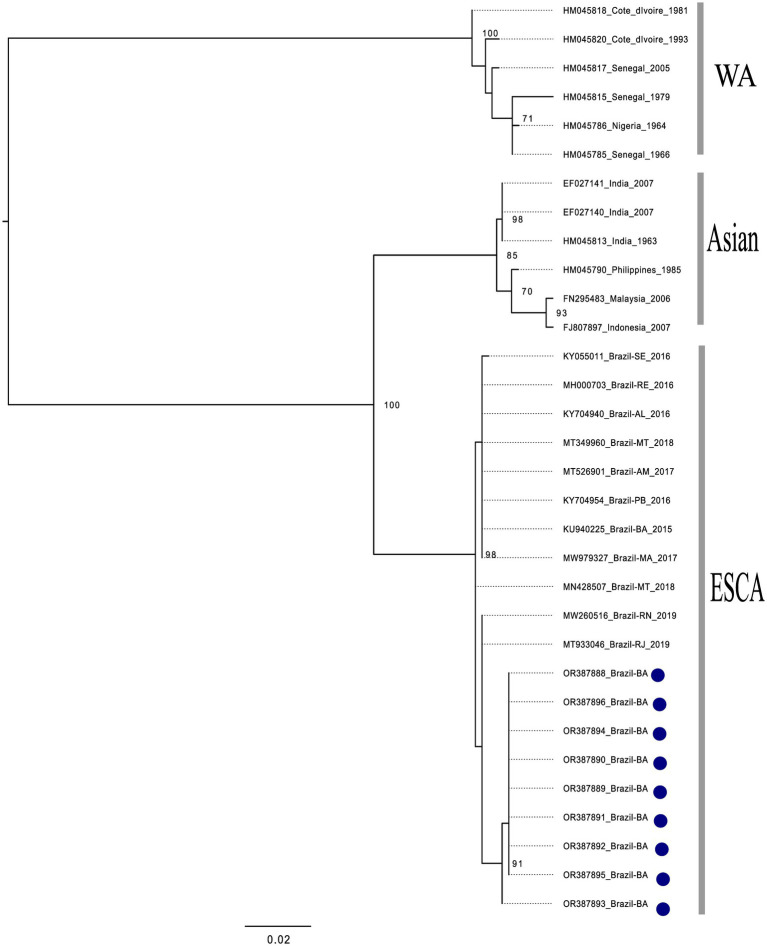
Figure 2.
Phylogenetic analysis of CHIKV, based on partial sequences of the E1-3’UTR (untranslated) region. The phylogenetic tree was constructed using the maximum likelihood method with a bootstrap of 1,000 replicates. The dataset consisted of 32 CHIKV nucleotide sequences with: West African genotype (n = 06), Asian genotype (n = 06), ECSA genotype (n = 11) and the sequences generated in this study (n = 9). All sequences retrieved from NCBI are identified in the format: accession number/country/year of isolation. Sequences generated in this study are identified according to its internal number and country of isolation (blue dots). The bootstrap values are very similar to the isolates from southeastern Brazil, being grouped with the isolates from Rio de Janeiro. It is thus inferred that these isolates belong to the ECSA lineage, the isolates from Bahia (2020–2021) form a monophyletic group with high support value, indicating that they had the same evolutionary origin.