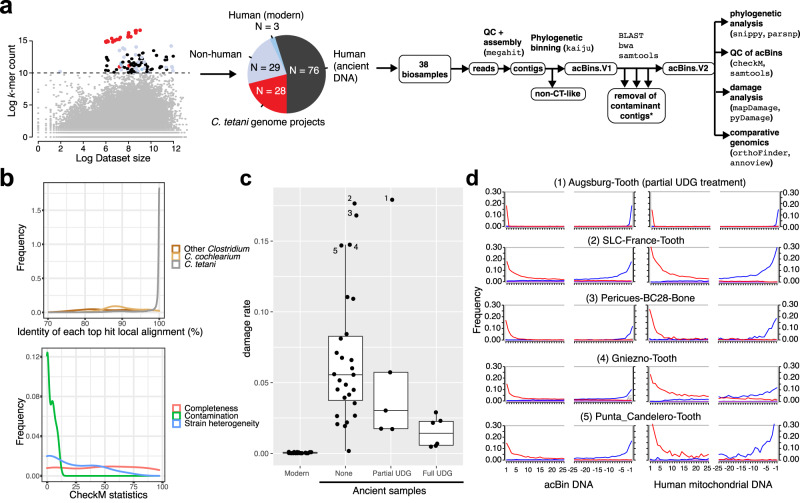
Fig. 1. Petabase-scale screen of the NCBI sequence read archive reveals C. tetani-related genomes in ancient human archeological samples.
a General bioinformatic workflow starting with the analysis of 43,620 samples from the NCBI sequence read archive. Each sample is depicted according to its C. tetani k-mer abundance (y axis) versus the natural log of the overall dataset size in megabases (x axis). A threshold was used to distinguish samples with high detected C. tetani DNA content, and these data points are colored by sample origin: modern C. tetani genomes (red), non-human (light blue), modern human (blue), ancient human (black). The pie chart displays a breakdown of identified SRA samples with a high abundance of C. tetani DNA signatures. The 38 aDNA samples predicted to contain C. tetani DNA were further analyzed as shown in the bioinformatic pipeline on the right. b Top—density plot of the percentage identities of all BLAST local alignments detected between acBins and reference genomes including C. tetani, C. cochlearium, and other Clostridium spp. Bottom—density plot of the checkM results for the 38 acBins including estimated completeness, contamination, and strain heterogeneity levels. Completeness and contamination levels are percentage values. c MapDamage damage rates (5’ C → T misincorporation frequency) for acBins (n = 38 biologically independent samples) subdivided by UDG treatment [none (n = 27), partial (n = 5), and full (n = 6)]. Also shown are the damage rates for modern C. tetani genomes (n = 21 biologically independent samples). The boxplots depict the lower quartile, median, and upper quartile of the data, with whiskers extending to 1.5 times the interquartile range (IQR) above the third quartile or below the first quartile. d Damage plots for the top five acBins with the highest damage rates, and corresponding mtDNA damage plots. Shown is the frequency of C → T (red) and G → A (blue) misincorporations at the first and last 25 bases of sequence fragments. Increased misincorporation frequency at the edges of reads is characteristic of ancient DNA. Source data for (a–d) are provided as a Source Data file.