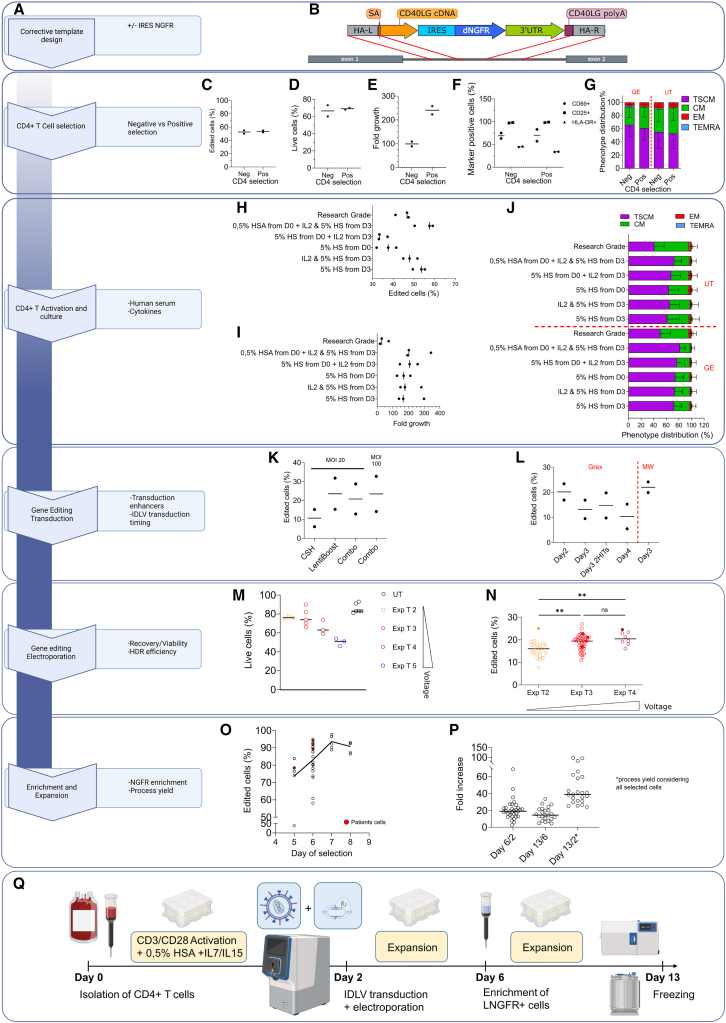
Figure 1.
Small- and medium-scale manufacturing of edited CD4+ T cells
(A) Flow chart with the decision points for manufacturing process development. For each step, corresponding panels are encircled in blue. (B) Gene editing strategy: the corrective cassette is integrated by homologous recombination (red lines) in the first intron of the CD40LG gene (dark gray). HA-L, homology arm left; SA, splice acceptor; HA-R, homology arm right. (C–E) Comparison of CD4+ selection by single-antibody-positive selection vs. negative selection by antibody cocktail in terms of editing efficiency (C), viability (D), proliferation rate (E), expression of activation markers (F), and phenotype (mean ± SD) (G). (H–J) Impact of human serum (HS) and IL-2 supplementation at different time points on HDR efficiency (H), cell growth (I), and phenotype (J) (mean ± SD), with AAV6 donor template. (K) Impact of transduction enhancers cyclosporine H (CsH)14 and Lentiboost on editing efficiency, alone or in combination (Combo) targeting the AAVS1 locus with a GFP reporter IDLV. (L) Impact of timing of transduction on editing efficiency, assessed by LNGFR expression in CD40LG locus with IDLV donor template (B). (M and N) Relative performance of different Maxcyte GTx electroporation protocols in terms of cell viability (M) and HDR efficiency (N). Kruskal-Wallis test with Dunn’s multiple comparisons. Exp T2-T5 correspond to increased voltage settings. Exp, expanded. (O) Immunomagnetic enrichment of LNGFR+ cells at different days after isolation and editing. (P) Fold growth at D6, D13, and cumulative growth of healthy donor (HD) cells. D6/D2 is the cell fold growth measured at day 6, 4 days after gene editing, before the cell selection step. D13/D6 is the fold growth obtained at the end of the process (from day 6 to day 13), D13/D2 is the total fold growth from the day of the electroporation to the end of the process considering the loss of cells after the selection step. (Q) Schematic representation of the optimized small- and medium-scale protocol. Cells were immunomagnetically selected from buffy coats or peripheral blood samples using CD4 microbeads and activated in G-Rex 24-6-6M on day 0; transduced with IDLV and electroporated with Cas9 RNP using MaxCyte GTx on day 2; on day 6 LNGFR+ cells were enriched and seeded for cell expansion; and, at day 13, cells were frozen. AAV6 donor was used in (C–J). IDLV donor was used in the remaining panels. Empty dots indicate HD cells; full dots indicate patient cells. Editing efficiency, phenotype, and activation markers were assessed by flow cytometry. UT, untreated; GE, gene edited; TSCM, T stem cell memory; CM, central memory; EM, effector memory; TEMRA, terminal differentiated; MOI, multiplicity of infection; MW, multiwell. Percentage of live cells was assessed by trypan blue exclusion.