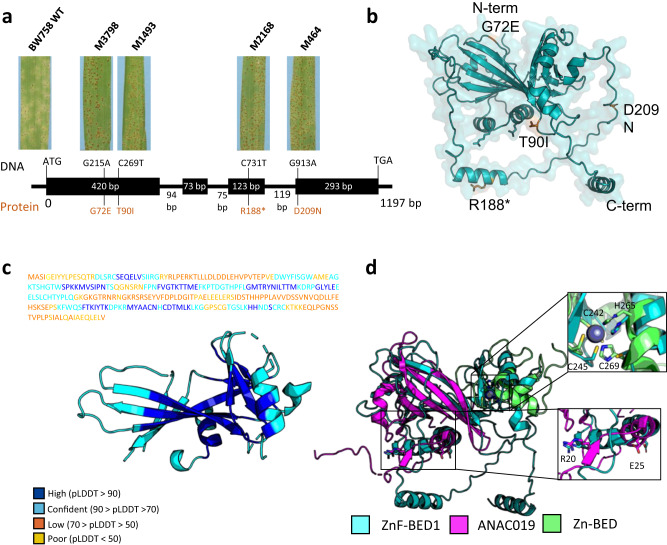
Fig. 2. ZnF-BED1 shares structural similarity with plant specific NAC domain-containing proteins.
a Schematical drawing of the gene structure of ZnF-BED1 consisting of 4 exons (black boxes) and 3 introns with respective length in bp. Phenotypic responses at the seedling stage 10 days after infection with Rph7-avirulent Puccinia hordei pathotype 5457 P+ of resistant wild type (WT) (BW758) and four independent sodium azide-induced mutants are shown for each of the mutant lines. Positions of four independent mutations with corresponding changes on DNA level (black) and protein level (orange) within the coding sequence indicating that the ZnF-BED1 gene was required for Rph7-mediated resistance. b Transparent surface representation of ZnF-BED1 highlighting the four non-synonymous amino acid changes identified in ZnF-BED1 mutant lines. Residues are shown in stick representation and coloured in orange. c AlphaFold2 prediction of ZnF-BED1 shown in cartoon representation. The predicted model is coloured by the per-residue confidence (pLDDT) score bands as depicted, with only confident regions (pLDDT > 70) shown. The sequence above the prediction is coloured according to confidence bands as represented. The full prediction can be found in Fig. S3a. d Structural superimposition of the AlphaFold2 prediction of ZnF-BED1 (teal) and top structural match from Dali ANAC019 NAC domain (PDB ID: 3SWP; magenta) shows similarity is limited to the N-terminus of ZnF-BED1, and superimposition of the top structural match from Dali for the C-terminal domain (residues 220-302), the C2H2-type zinc-finger domain of human zinc-finger BED domain-containing protein 2 (PDB ID: 2DJR, green). Insert shows residues involved in Zn co-ordination in 2DJR, and dimerisation in 3SWP, and corresponding putative residues that may be involved in ZnF-BED1. Residues are labelled according to ZnF-BED1.