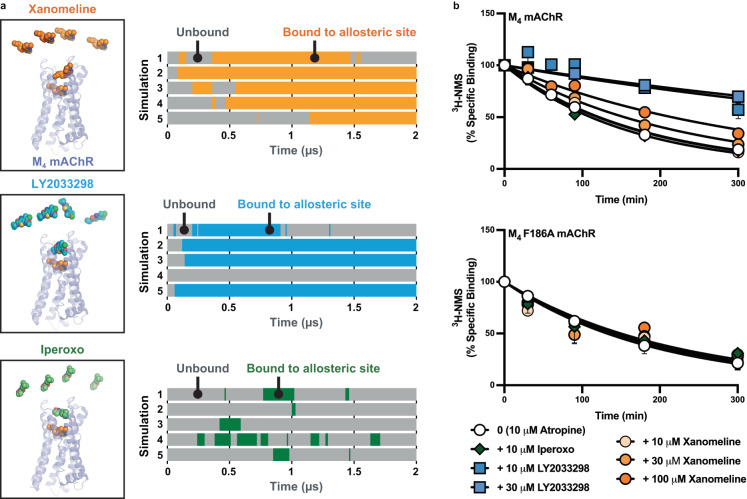
Fig. 3. Computational and pharmacological validation of xanomeline in the allosteric binding site.
a Molecular dynamics simulations reveal that xanomeline spontaneously binds to the M4 mAChR allosteric site for a similar fraction of time as the prototypical PAM, LY2033298, and for a substantially longer fraction of time than the orthosteric agonist, iperoxo. Simulations were initiated with a xanomeline molecule bound in the orthosteric site and with the free ligands in solution—either xanomeline, LY2033298 or iperoxo—all being at the same concentration. Each horizontal bar represents an independent simulation and indicates the amount of time that the allosteric site is vacant (grey) or ligand-bound (non-grey). b [3H]-N-methylscopolamine ([3H]-NMS) dissociation via isotopic dilution with 10 µM atropine alone (0), or in the presence (+), of xanomeline, LY2033298, or iperoxo, at the M4 mAChR wild type and M4 F186ECL2A mutant. Data points represent the mean ± S.E.M. of three to nine individual experiments performed in duplicate. M4 mAChR wild type; 10 µM atropine alone n = 14, + 10 µM iperoxo n = 5, + 30 µM LY2033298 n = 7, + 10 µM LY2033298 n = 4, + 10 µM xanomeline n = 6, + 30 µM xanomeline n = 8, + 100 µM xanomeline n = 13. M4 F186ECL2A; 10 µM atropine alone n = 4, + 10 µM iperoxo & + 30 µM LY2033298 & + 30 µM xanomeline & + 100 µM xanomeline n = 3. A one-phase exponential decay model was fit to the data.