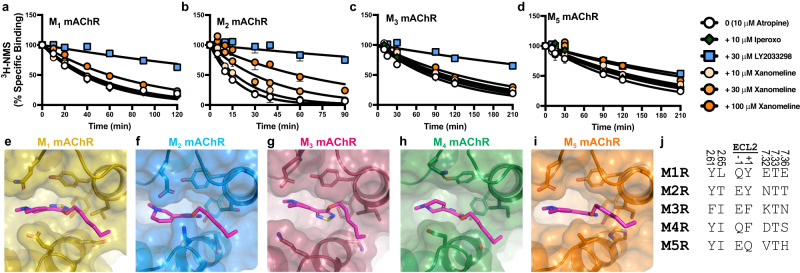
Fig. 4. Xanomeline binds allosterically at all mAChR subtypes.
a–d [3H]-N-methylscopolamine ([3H]-NMS) dissociation via isotopic dilution with 10 µM atropine alone (0), or in the presence (+), of xanomeline, LY2033298, or iperoxo, at the a M1, b M2, c M3 or d M5 mAChRs (M1R-M5R). Data points represent the mean ± S.E.M. of four to twelve individual experiments performed in duplicate. M1 mAChR; 10 µM atropine alone & + 10 µM iperoxo & + 10 µM xanomeline & + 30 µM xanomeline & + 100 µM xanomeline n = 6, + 30 µM LY2033298 n = 3. M2 mAChR; 10 µM atropine alone n = 10, + 10 µM iperoxo & + 30 µM LY2033298 & + 10 µM xanomeline & + 30 µM xanomeline & + 100 µM xanomeline n = 6. M3 mAChR; 10 µM atropine alone & + 10 µM iperoxo & + 10 µM xanomeline & + 30 µM xanomeline & + 100 µM xanomeline n = 7, + 30 µM LY2033298 n = 6. M5 mAChR; 10 µM atropine alone & + 10 µM iperoxo & + 10 µM xanomeline & + 30 µM xanomeline & + 100 µM xanomeline n = 5, + 30 µM LY2033298 n = 4. A one-phase exponential decay model was fit to the data. The allosteric binding site of each mAChR subtype can accommodate the binding of xanomeline, as shown by energy minimization of xanomeline in the allosteric site of the e M1 (PDB: 6OIJ), f M2 (PDB: 6OIK), g M3, h M4, or i M5 mAChRs. j Sequence comparison of residues that contact xanomeline in the allosteric site (≤4 Å) across all five mAChR subtypes. Residues in TM2 and TM7 are labelled with the Ballesteros and Weinstein scheme for class A GPCRs and residues in ECL2 are numbered according to their relative position of a conserved cysteine residue (C185 at the M4 mAChR).