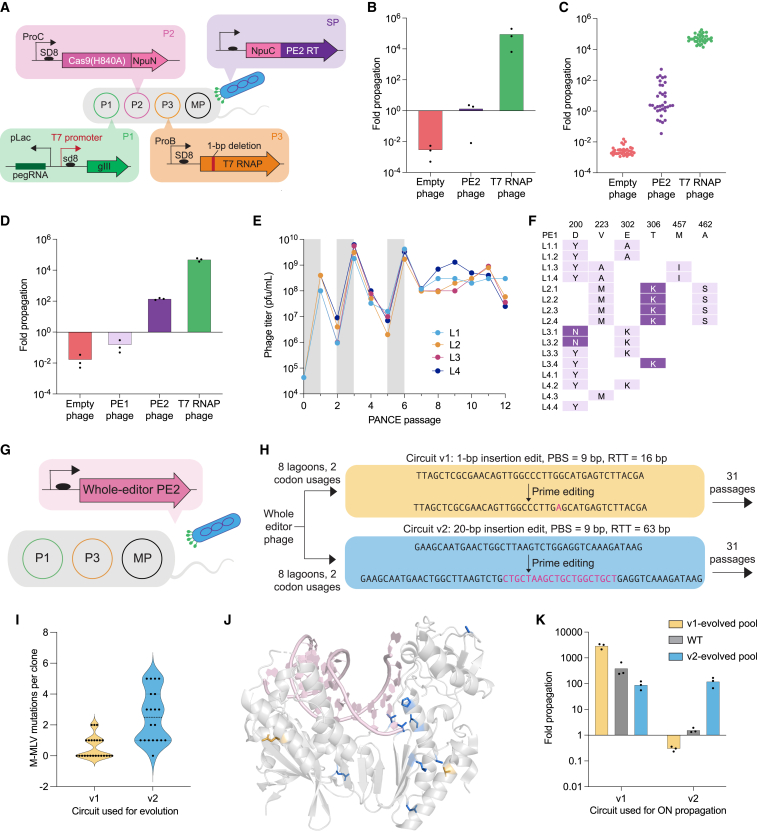
Figure 2.
Development and validation of a prime editing PACE selection
(A) Schematic of PE-PACE selection circuit. Upon infection of E. coli by selection phage (blue), the NpuN intein and NpuC intein (pink) mediate reconstitution of the PE2 prime editor (purple and pink), which engages a pegRNA (dark green) and corrects a frameshift in T7 RNAP (orange) via PE. Functional T7 RNAP then transcribes gIII (light green), which enables SP propagation.
(B) Phage replication levels from overnight propagation of empty phage (red), NpuC-PE2-RT phage (purple), and T7-RNAP phage (green) in PE-PACE host cells before pegRNA optimization.
(C) Screen of pegRNAs for the v1 PE-PACE circuit. Overnight propagation values of empty phage (red), NpuC-PE2-RT phage (purple), and T7-RNAP phage (green) are shown. Each point reflects the mean value of n = 3 independent biological replicates for a different pegRNA. Individual replicates are shown in Figure S2C.
(D) Overnight propagation of empty phage (red), NpuC-PE1-RT phage (light purple), NpuC-PE2-RT phage (dark purple), and T7-RNAP phage (green) in the v1 pegRNA-optimized circuit.
(E) PANCE titers for the evolution of NpuC-PE1-RT phage. Gray shading indicates a passage of evolutionary drift, in which phage were supplied gIII in the absence of selection. Titers of four replicate lagoons are shown.
(F) Mutation table for NpuC-PE1-RT phage surviving v1 PANCE. Four clones per lagoon (L1-L4, with clones ordered by lagoon) were sequenced. Light purple denotes conserved mutations. Dark purple denotes conserved mutations also present in the previously engineered PE2 RT1.
(G) Schematic of the PE-PACE selection for evolution of the whole prime editor, including the Cas9 domain. The P1 plasmid (green) and P3 plasmid (orange) are identical to those used in Figure 2A.
(H) PANCE experiment to compare the outcome of selection on v1 and v2 selection circuits. Replicate lagoons were evolved on each (v1, yellow and v2, blue) selection circuit. After 31 passages, clones from each selection were sequenced, and the resulting mutations were compared to generate (I-K).
(I) Violin plots showing the number of mutations per clone for the M-MLV domain of whole-editor phage evolved with either the v1 (yellow) or v2 (blue) circuit. Data are shown as individual values, with one dot representing one sequenced phage. The mean value is shown as a dotted line.
(J) Predicted positions of mutated residues in M-MLV from v1 (yellow) or v2 (blue) PANCE. The structure is from the highly homologous XMRV (PDB: 4HKQ).
(K) Overnight propagation of pools of wild-type RT and evolved RT phage on their cognate or noncognate host-cell selection strains. Phage were from PANCE on the v1 circuit (yellow bars), from PANCE on the v2 circuit (blue bars), or wild-type-PE2 phage (gray bars). Propagation was then measured in the v1 circuit (left) or the v2 circuit (right). Bars reflect the mean of n = 3 independent replicates, and dots show individual replicate values (B, D, K). See also Figure S2.