Figure 4.
Development of dual-AAV compatible RT variants for installing long, complex edits
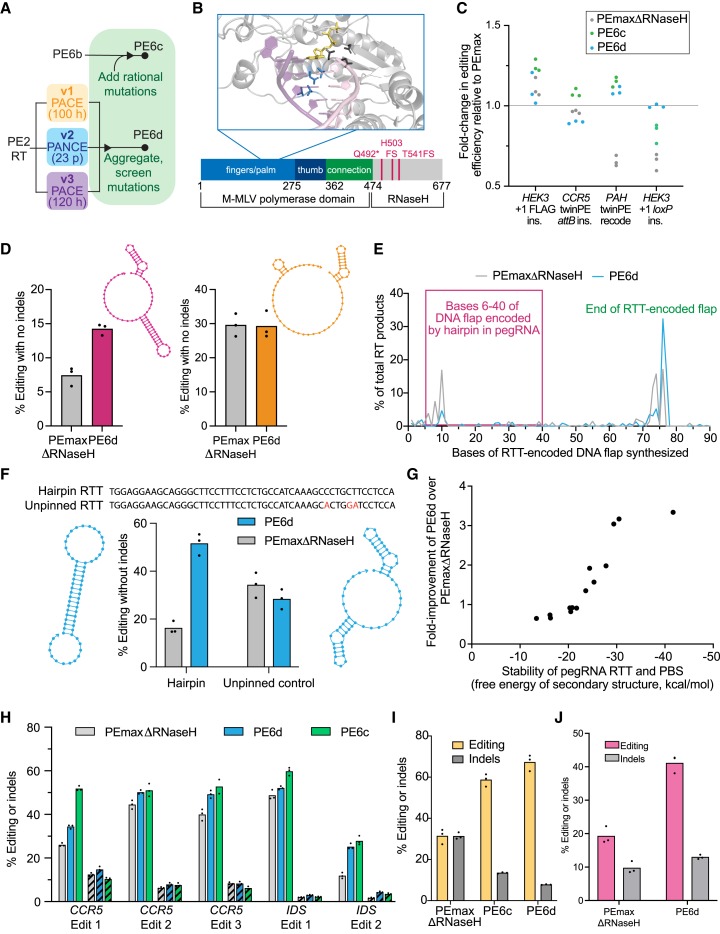
(A) Summary of evolution and engineering campaigns used to generate PE6c and PE6d.
(B) Conserved mutations from M-MLV RT evolution. The structure of XMRV RT (PDB: 4HKQ), which is highly homologous to M-MLV shows PACE-evolved residues (blue) lie close to the enzyme active site (dark gray) and DNA/RNA duplex substrate (pink/purple). An incoming dNTP, modeled by alignment with PDB: 5TXP, is shown in yellow. Below, pink lines indicate locations in the M-MLV RT at which PACE-evolved mutations truncated the protein.
(C) Fold-change in editing efficiency relative to PEmax for PEmaxΔRNaseH, PE6c, and PE6d in HEK293T cells. Individual replicates are plotted, with n = 3 biological replicates per edit.
(D) Editing efficiencies of PEmaxΔRNaseH and PE6d at the HEK3 +1 loxP insertion edit (pink) and the HEK3 +1 FLAG insertion edit (orange) in HEK293T cells. The NUPACK-predicted structures of the RTT and PBS extensions for each edit is shown.
(E) Results of a TdT assay on the HEK3 +1 loxP insertion edit in HEK293T cells. The y axis indicates the percentage of total RT products of a given length, and the x axis represents the length of the product in base pairs. PEmaxΔRNaseH is shown in gray, and PE6d is shown in blue. The lines are mean values from n = 3 biological replicates. The pink box indicates DNA bases templated by the structured portions of the pegRNA.
(F) Editing efficiencies of PEmaxΔRNaseH (gray) and PE6d (blue) at an example engineered hairpin edit and its corresponding unpinned control in HEK293T cells. The sequence of the RTT is shown, with point mutations in the unpinned control shown in red. The NUPACK-predicted structures of the RTT and PBS extensions for each edit is shown.
(G) Relationship between pegRNA RTT/PBS secondary structure and PE6d improvements. The y axis reflects the fold-improvement of PE6d over PEmaxΔRNaseH. The x axis is the absolute value of the free energy of pegRNA folding as measured by NUPACK. Each dot represents one edit in HEK293T cells that was calculated from the mean values from n = 3 biological replicates. See Figure S4D for individual editing values and edit identities.
(H) Comparison of evolved and engineered RTs to PEmaxΔRNaseH at typical twinPE edits in HEK293T cells. Solid bars indicate editing efficiency. Striped bars indicate indels.
(I) TwinPE-mediated insertion of the 38-bp attB sequence into the Rosa26 locus in N2a cells. Indel-free editing is shown in yellow, and indels are shown in gray.
(J) PE-mediated insertion of a 42-bp sequence containing loxP into the Dnmt1 locus in N2a cells. Indel-free editing is shown in yellow, and indels are shown in gray.
For D, F, and H-J, bars reflect the mean of n = 3 independent replicates. Dots show individual replicate values. See also Figure S4.