Figure 6.
Evolution and engineering of improved Cas9 domains for prime editing, and summary of PE6 recommended use cases
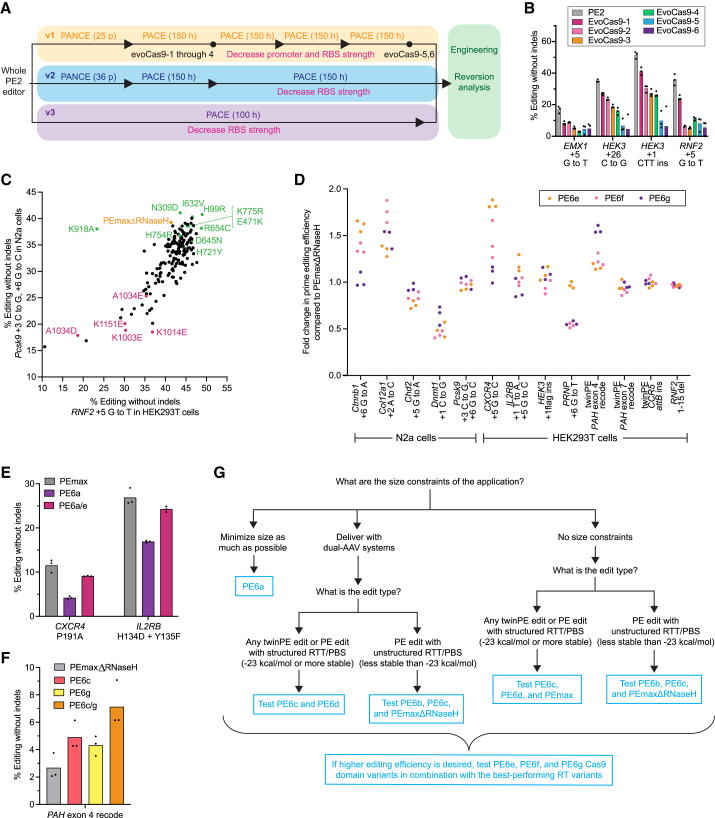
(A) Summary of evolution campaigns for whole PE2 phage in the v1 (yellow), v2 (blue), and v3 (purple) circuits. Green shading indicates reversion analysis. PANCE passages (p) or hours of PACE (h) are in parentheses. Arrowheads indicate increases in selection stringency. Mutants characterized in mammalian cells are denoted with a dot and labeled. Additional increases in stringency are in pink.
(B) Evaluation of PACE-evolved clones in HEK293T cells. EvoCas9-1 through evoCas9-4 were isolated from low-stringency evolution. EvoCas9-5 and evoCas9-6 were isolated from high-stringency evolution.
(C) Assessment of individual Cas9 mutations on prime editing efficiency at two test sites. The y axis shows editing efficiency at the Pcsk9 +3 C to G / +6 G to C edit in N2a cells. The x axis shows editing efficiency for the RNF2 +5 G to T edit in HEK293T cells. Mutants incorporated into final Cas9 variants are shown in green. Mutants previously shown to, or structurally predicted to, decrease Cas9 binding are shown in maroon. PEmaxΔRNaseH is shown in orange.
(D) Comparison of combined Cas9 mutants to PEmaxΔRNaseH in HEK293T cells and N2a cells. Editing efficiencies of variants are normalized to the editing efficiency generated by PEmaxΔRNaseH. Individual replicates are plotted, with n = 3 biological replicates per edit.
(E) Comparison of PEmax, PE6a, and PE6a/e at two sites in HEK293T cells.
(F) Comparison of PEmaxΔRNaseH, PE6c, and PE6g in HEK293T cells.
(G) Decision tree for selecting a PE6 variant. For secondary structure stability predictions, we recommend the NUPACK prediction tool38 with the RTT/PBS sequence as the input.
For B, E, and F, bars reflect the mean of n = 3 independent replicates. Dots show individual replicate values. See also Figure S6.