Figure S1.
Characterization and engineering of reverse transcriptase enzymes for prime editing, related to Figure 1
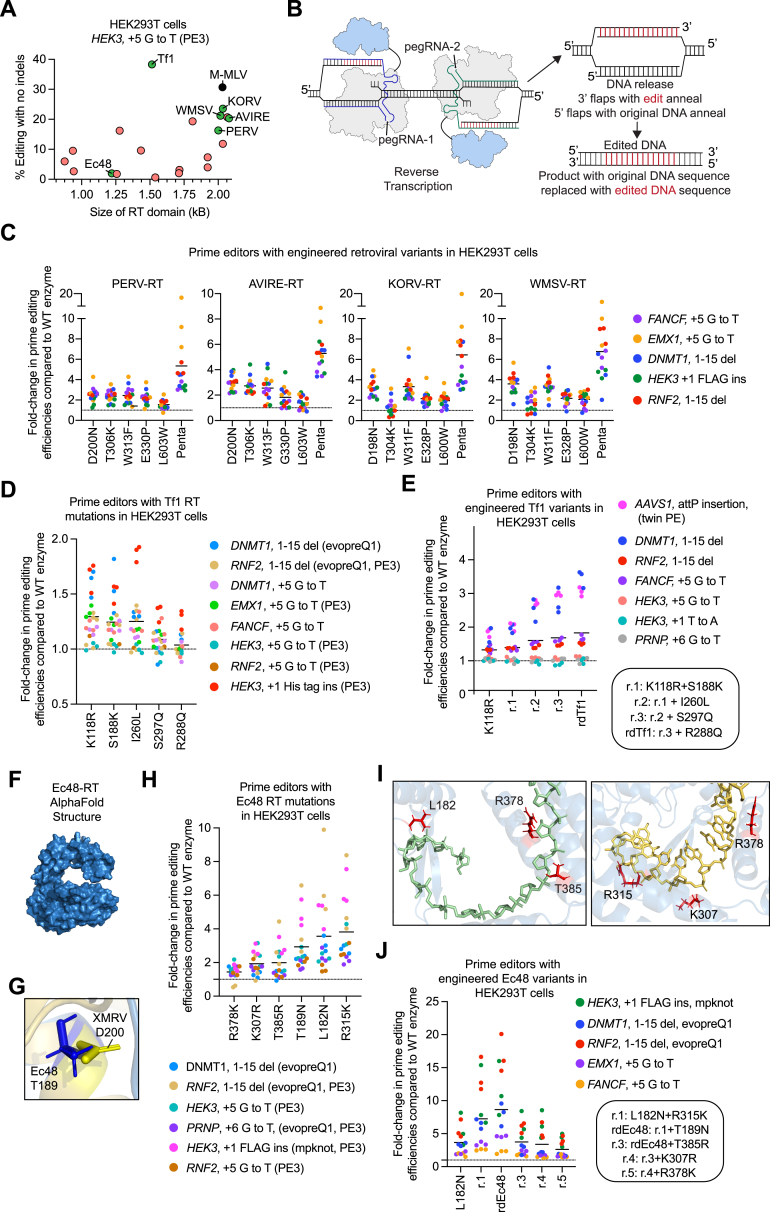
(A) Native small RT enzymes demonstrate poor activity in the prime editing system (HEK293T cells, HEK3 +5 G to T edit). RT enzymes engineered in Figure 1 are highlighted in green, and the wild-type M-MLV RT used in the PE1 system is highlighted in black. All other enzymes are in red. Dots reflect the mean of n = 3 independent replicates. Of these enzymes that can support detectable mammalian PE activity, 11 are closely related to the M-MLV RT and are encoded by retroviruses, two are encoded by LTR retrotransposons, and seven are bacterial RTs from group-II introns, retrons, or CRISPR-Cas associated systems.
(B) Overview of twinPE. The prime editor protein (gray and blue) uses two pegRNAs (dark blue and teal) to target opposite strands of DNA. The prime editor generates two 3’ flaps (red) that are complementary to each other. After these newly synthesized 3’ flaps anneal and the original DNA sequence in the 5′ flaps is degraded, the edited sequence in the flaps is permanently installed at the target DNA site.
(C) Incorporation of each of the five mutations analogous to those in PE2 (D200N, T306K, W313F, T330P, and L603W) improves the activity of four retroviral RT enzymes in HEK293T cells. PERV = porcine endogenous retrovirus RT, AVIRE = avian reticuloendotheliosis virus RT, KORV = koala retrovirus RT and WMSV = woolly monkey sarcoma virus RT. Combining all five mutations together (Penta) further improves the activity of each enzyme. All values from n = 3 independent replicates are shown. Horizontal bars show the mean value.
(D) Structure-guided rational engineering of the Tf1 RT identifies five mutations that improve prime editing in HEK293T cells. The solved structure of the Tf1 RT homolog, Ty3 RT, was used to predict mutations that could increase contacts of the RT with its DNA-RNA substrate (PDB: 4OL8). All values from n = 3 independent replicates are shown. Horizontal bars show the mean value across all sites and replicates.
(E) Combining all mutations identified from structure-guided rational engineering improves the activity of the Tf1 RT prime editor in HEK293T cells. The final rationally designed Tf1 variant (rdTf1) is a combination of five mutations: K118R, S188K, I260L, R288Q and S297Q. All values from n = 3 independent replicates are shown. Horizontal bars show the mean value.
(F) AlphaFold-predicted structure of the Ec48 RT enzyme. The predicted structure aligns well with the RT from the xenotropic murine leukemia virus-related virus (XMRV, PDB: 4HKQ), a close relative of the M-MLV RT.70
(G) Aligning the AlphaFold-predicted structure of the Ec48 RT (blue) with the RT from xenotropic murine leukemia virus-related virus (XMRV, PDB: 4HKQ, yellow), a close relative of the M-MLV RT, suggests that the residue analogous to the D200 residue in M-MLV RT is the T189 residue in Ec48 RT.
(H) Structure-guided rational engineering of the Ec48 RT identifies six mutations that improve prime editing in HEK293T cells. An AlphaFold-generated predicted structure of the Ec48 RT was overlayed with the structure of the RT from the xenotropic murine leukemia virus-related virus (XMRV) (PDB: 4HKQ) to perform structure-guided mutagenesis. All values from n = 3 independent replicates are shown. Horizontal bars show the mean value.
(I) Positions of residues (red) proximal to the substrate that were mutated to improve the activity of the Ec48 RT prime editor. Residues are mapped onto the predicted AlphaFold structure of the Ec48 RT aligned with the solved substrate of the XMRV RT (PDB: 4HKQ). L182 and T385 are proximal to the DNA substrate (green), R315 and K307 are proximal to the RNA substrate (yellow) and R378 is proximal to both the DNA and RNA rate.
(J) Combining the top three mutations identified from structure-guided engineering improves the activity of the Ec48 RT prime editor in HEK293T cells. The final rationally designed Ec48 RT variant (rdEc48) contains three mutations: L182N, T189N and R315K. All values from n = 3 independent replicates are shown. Horizontal bars show the mean value.