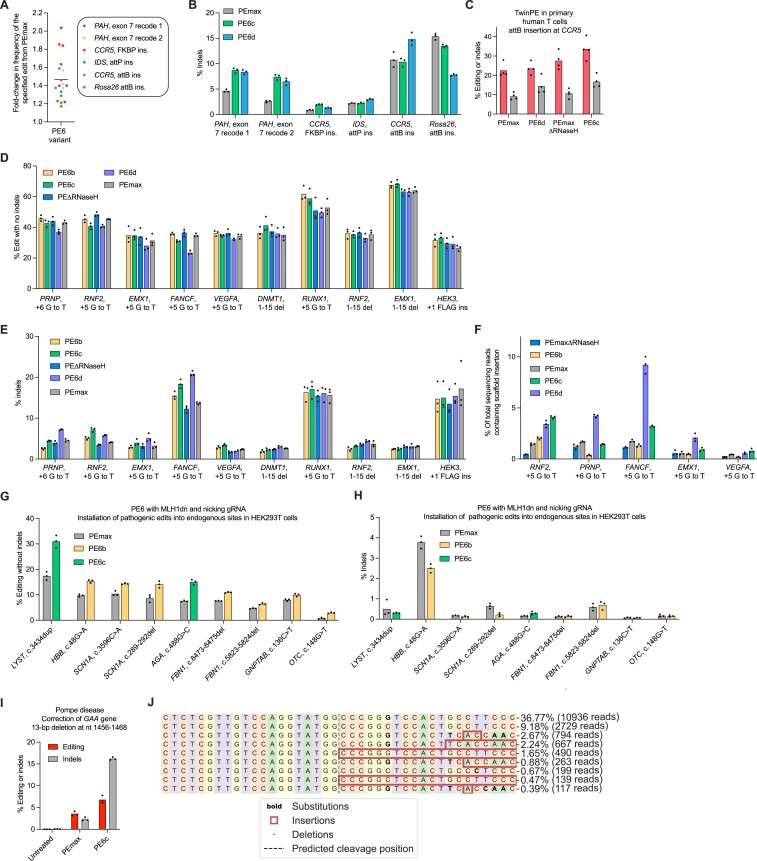
Figure S5.
Comparison of PE6 variants with PEmax, related to Figure 5
(A) Prime editing efficiencies of the best performing PE6 variant (either PE6c or PE6d) normalized to the editing efficiency of PEmax at sites tested in Figure 5A. All values from n = 3 independent replicates are shown. Editing was performed in HEK293T cells. The horizontal bar shows the mean value.
(B) Indel frequencies of PEmax, PE6c, and PE6d at edits tested in Figure 5A. This data was used for Figure 5B. Bars reflect the mean of three independent replicates. Editing was performed in HEK293T cells. Dots show individual replicate values.
(C) Screening PE6 variants for insertion of attB into the CCR5 locus in primary human T cells. Bars reflect the mean of n = 4 independent replicates for editing (red) and indels (gray). Dots show individual replicate values.
(D) Absolute prime editing efficiencies of PE6 variants, PEmaxΔRNaseH, and PEmax in HEK293T cells used to plot data for Figures 5D and 5E. Prime editing efficiencies used are the frequency of the intended prime editing outcome with no indels or other changes at the target site. Bars reflect the mean of three independent replicates. Dots show individual replicate values.
(E) Indel frequencies of PE6 variants, PEmaxΔRNaseH, and PEmax in HEK293T cells used to plot data for Figures 5D and 5E. Bars reflect the mean of three independent replicates. Dots show individual replicate values.
(F) Percentage of sequencing reads containing a pegRNA scaffold insertion after prime editing using PE6 variants, PEmaxΔRNaseH, and PEmax in HEK293T cells. These reads contribute to the total indel frequency. Bars reflect the mean of n = 3 independent replicates. Dots show individual replicate values.
(G) Prime editing efficiencies for edits where PE6b or PE6c outperformed PEmax using a nicking gRNA. Bars reflect the mean of n = 3 independent replicates. Dots show individual replicate values. Prime editing efficiencies used are the frequency of the intended prime editing outcome with no indels or other changes at the target site in HEK293T cells.
(H) Indel frequencies of PE6 variant and PEmax at sites shown in Figure 5F in HEK293T cells. Bars reflect the mean of n = 3 independent replicates. Dots show individual replicate values.
(I) Correction of mutation implicated in Pompe disease in patient-derived fibroblast using PE6c and PEmax. Bars reflect the mean of n = 3 independent replicates for editing (red) and indels (gray). Dots show individual replicate values.
(J) Distribution of editing outcomes after correction of the pathogenic mutation implicated in Pompe disease in patient-derived fibroblasts using PE6c. The patient was heterozygous. Indel genotypes are shown. Interestingly, many of the indels detected at this site did not contain the silent PAM edit encoded by the pegRNA, suggesting those indels were not RT-templated products.