Figure 5.
Altered gene expression in excitatory neuronal nuclei during seizures
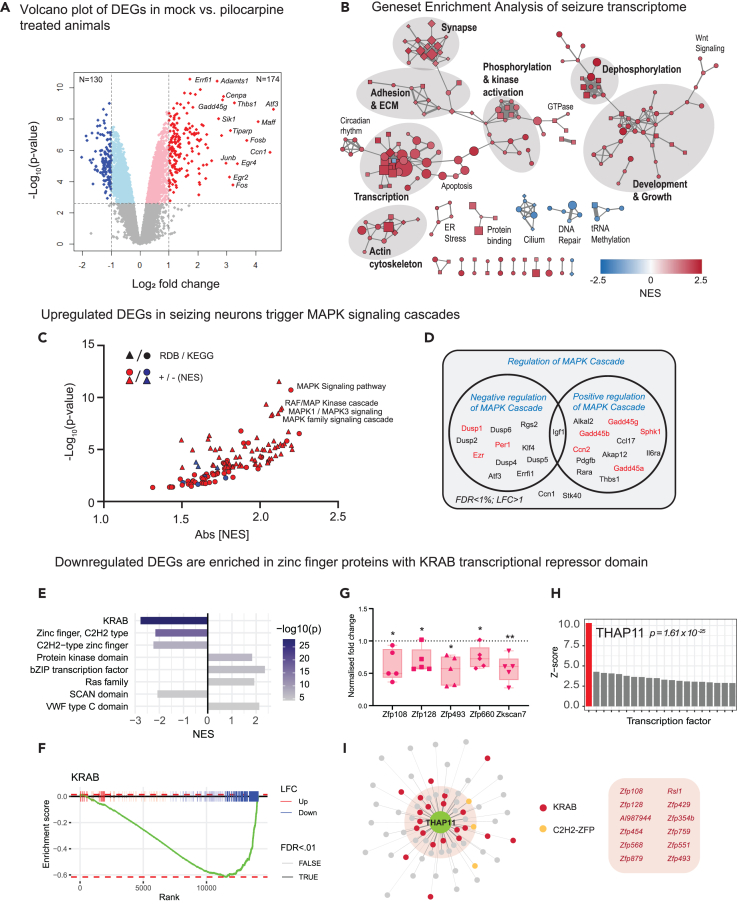
(A) Volcano plot showing differential expression of protein-coding genes in mock-vs. pilocarpine-treated animals at 100 min. Thresholds for differential expression are indicated at false discovery rate (FDR) < 1% and log2 fold change (LFC) > ±1, at which N = 130 genes are downregulated and N = 174 are upregulated.
(B) Gene set enrichment analysis (GSEA) of the seizure-induced transcriptome (p < 0.05). Each node corresponds to an enriched GO term (circle: Biological Process (BP), square: Molecular Function (MF), diamond: Cellular Component (CC)) with node color representing nucleotide enrichment score (NES) and larger nodes having lower p values. Edges indicate gene set overlap, with thicker edges representing greater overlap. Clusters of related nodes were labeled manually. Disconnected nodes are not shown.
(C) GSEA of ReactomeDB (RDB) and KEGG pathways.
(D) DEGs involved in the regulation of MAPK cascade according to GO annotations. Genes in red regulate stress-activated MAPK signaling.
(E) GSEA of PFAM protein families.
(F) GSEA enrichment plot showing KRAB box genes ranked by LFC in the seizure vs. mock transcriptome (padj <0.01). Up and downregulated genes and DEGs are indicated. Enrichment score is plotted for each point in the ranked list, showing enriched downregulation of KRAB genes.
(G) qPCR validation of zinc-finger protein genes Zfp108, Zfp128, Zfp493, Zfp660, and Zkscan7 (N = 5 pairs of mock v. pilocarpine mice). Relative gene expression levels determined by normalizing against average ΔCt values for all mock-injected animals. All statistics were performed using one-sample t test with plots showing means ± SEM, p < 0.05 (∗), p < 0.01 (∗∗).
(H) Analysis of enriched transcription factor binding sites among downregulated DEGs using Pscan. Z-scores indicating target site enrichment for top 20 transcription factors.
(I) THAP11 target genes with maximum binding scores above background average. Top targets containing KRAB domains are listed.