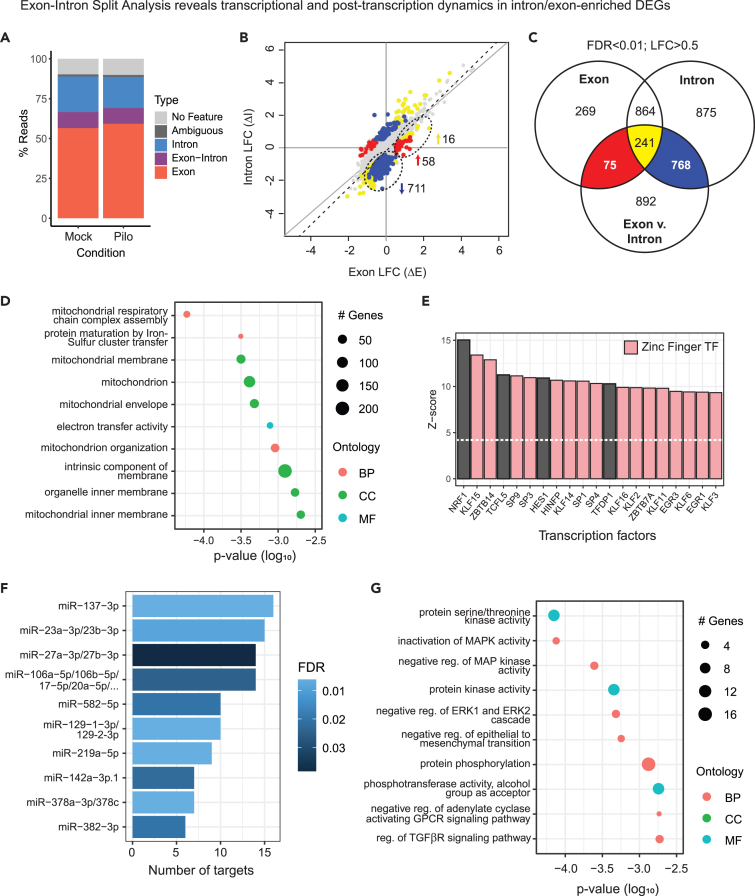
Figure 6.
Differential gene expression at intron and exon levels reveals transcriptional dynamics and putative post-transcriptional regulation
(A) Percentage of reads uniquely aligned to exons, introns, intergenic regions, and ribosomal RNA (rRNA), aligned ambiguously or unaligned.
(B) Log fold changes (LFCs) in gene expression between mock and seizure in introns (ΔI) versus exons (ΔE) for protein-coding genes with >1 log2CPM at both levels (N = 11,389). Colored subsets of genes have significantly different ΔI vs. ΔE and are differentially expressed between mock and seizure in exons (red), introns (blue), or both (yellow). Subsets of interest are circled. Linear regression is indicated by the dashed line.
(C) Venn diagram of genes differentially expressed between mock and seizure in exons and introns, and genes with significantly different ΔI vs. ΔE, corresponding to subsets indicated on LFC plot.
(D) Top 10 terms in GO analysis of intron-specific downregulated DEGs showing number of significant genes in enriched terms and log10 p value (pink: Biological Process (BP), cyan: Molecular Function (MF), green: Cellular Component (CC)).
(E) Enriched transcription factor-binding sites for intron-specific downregulated DEGs using Pscan. Z-scores indicate target site enrichment for top 20 transcription factors (TFs), of which 16 are zinc-finger TFs.
(F) Enrichment of miRNA target sites in TargetScan database using MIENTURNET for DEGs with significantly greater ΔE than ΔI. Number of miRNA target genes is plotted against false discovery rate (FDR) for top 10 miRNAs by FDR.
(G) Top 10 terms in GO analysis of DEGs with significantly greater ΔE than ΔI (pink: BP, cyan: MF, green: CC).