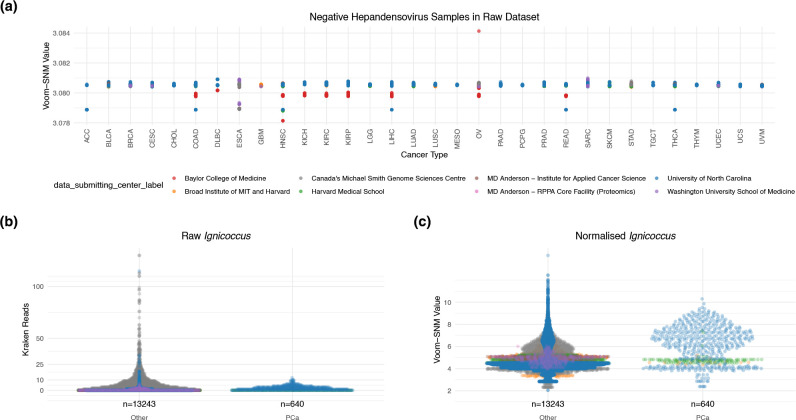
Fig. 1.
(a) Voom-SNM normalized TCGA samples (n=17 624) that were negative for crustacean virus hepandensovirus with zero classified reads in the original Kraken dataset with the most stringent decontamination approach. One sample contained two sequencing reads for Hepandensovirus, which has been omitted from this figure to illustrate inappropriate variation introduced by SNM. The colour of each point indicates the centre where the sample was sequenced and from where the resulting data were submitted [University of North Carolina, Harvard Medical School, Canada’s Michael Smith Genome Sciences Centre, Broat Institute MIT and Harvard, Baylor College of Medicine, Washington University School of Medicine, MD Anderson – Institute for Applied Cancer Science, Johns Hopkins/University of Southern California, MD Anderson RPPA Core Facility (Proteomics)]. The x-axis demonstrates cancer types using TCGA abbreviations as in Poore et al. [1]. This is a prominent concern, especially given how closely linked sequencing centre and disease type are (Table S3). Raw (b) and Voom-SNM normalized (c) Ignicoccus values, which was deemed the most important feature for predicting prostate cancer (PCa) from all other cancer types (n=13 883 primary tumours). Median values are as follows: Kraken raw other 0, Kraken raw PCa 1, normalized other 4.49, normalized PCa 5.82. In both the raw and normalized cases, the distributions are significantly different (Wilcox signed rank-sum test P<2.2×10–16).