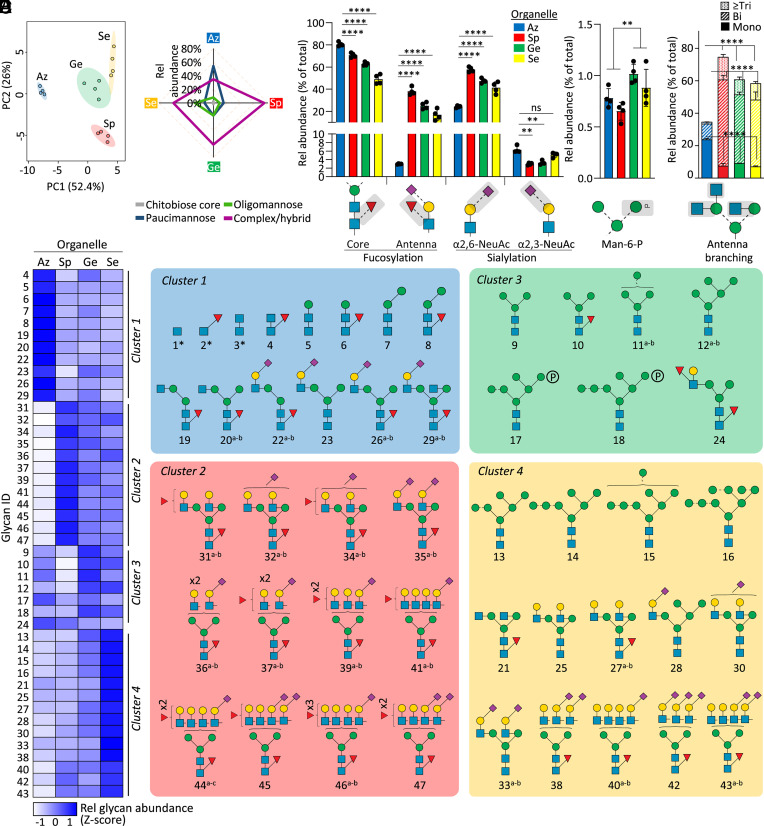
Fig. 2.
Organelle-specific N-glycome signatures in resting neutrophils. (A) PCA of quantitative N-glycomics data of the four cytosolic compartments (Az, Sp, Ge, and Se) isolated from resting neutrophils. (B) N-glycan class distribution across compartments. Key glycofeatures across organelles including (C) core/antenna fucosylation and α2,3-/α2,6-sialylation, (D) mannose-6-phosphate (M6P) modifications, and (E) antennary branching patterns (relative abundance of each glycofeature against the total N-glycome). For panel B−E, data plotted as mean and SD (n = 4 biological replicates, SD left out in panel B for simplicity, ANOVA test, **P < 0.01, ****P < 0.0001, ns, not significant). Organelle-specific differences were also observed for fucosylation, sialylation, M6P, and antennary branching when measured as a proportion of identified structures having the potential to be modified by the specific glycofeature rather than against the total N-glycome, an informative measure when exploring biosynthetic relationships associated with observed N-glycome differences (SI Appendix, Fig. S1 A and B). (F, Left) Unsupervised clustering (see SI Appendix, Fig. S1C for dendrogram) and heatmap of N-glycomics data illustrating four major clusters representing the neutrophil organelles. Data plotted as average of four biological replicates in each compartment at the composition level (relative abundances of N-glycan isomers were summed) and Z-score transformed. (Right) N-glycan structures contributing to each cluster. The most common structure is depicted for glycan compositions exhibiting multiple isomers. *chitobiose core-type N-glycans were prevalently observed at the glycopeptide level and, thus, for balance, were added to this map. See Fig. 1 for key. See Dataset S2 for glycan identifiers consistently used throughout the study.