Figure 4. Novel resistance mutations in SUGP1.
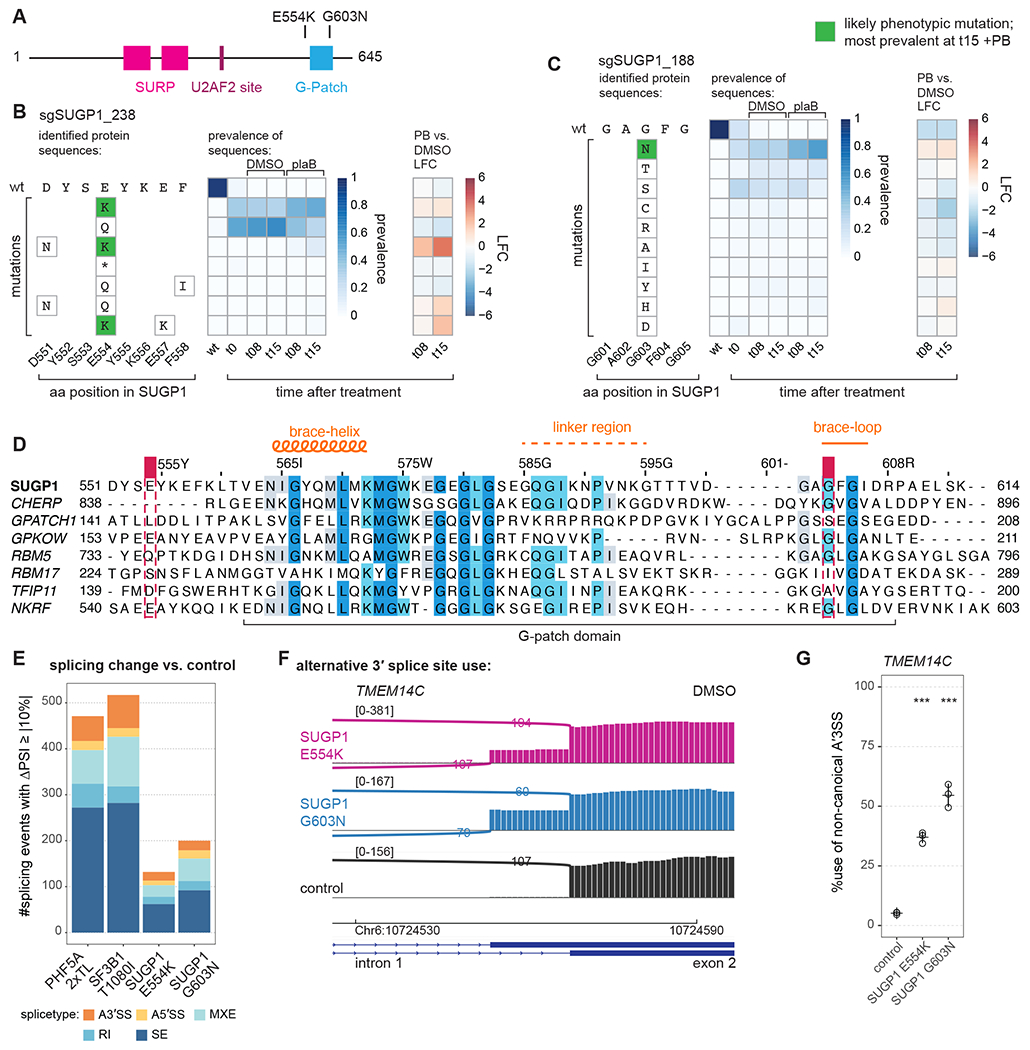
(A) Schematic of SUGP1 and its domains and motifs.
(B) Editing outcome for sgSUGP1_238.
(C) Editing outcome for sgSUGP1_188.
(D) Sequence alignment of all human G-patch motifs involved in splicing with the NKRF G-patch motif included as a reference. Shaded residues: more than 30% identity. Positions of mutants identified in screen are indicated. Alignment by JalView.
(E) Identified splicing changes for mutant vs. control cell lines. Numbers are shown for junctions identified by rMATS with FDR > 0.01 and |ΔPSI| > 10 (ΔPSI: PSI of mutant sample -PSI of control sample, where PSI: percent spliced in). RNA-seq data of total, polyA-selected RNA from three independent clonal cell lines treated for 3 h with DMSO. (A3’SS: alternative 3’ splice site use; A5’SS: alternative 5’ splice site use; MXE: mutually exclusive exon; RI: retained intron; SE: skipped exon.)
(F) Sashimi plot for alternative 3’ splice site usage in TMEM14C exon 2 (DMSO). Representative traces for a single clonal cell line are shown.
(G) RT-PCR and quantification for alternative 3’ splice site usage for TMEM14C exon 2 (DMSO). Statistical analysis for RT-PCR: one-way ANOVA with Dunnett’s for multiple comparison (two-sided, with control as reference) was performed with R and package multcomp; * p < 0.05, ** p < 0.01, and *** p < 0.001; all with n = 3.
