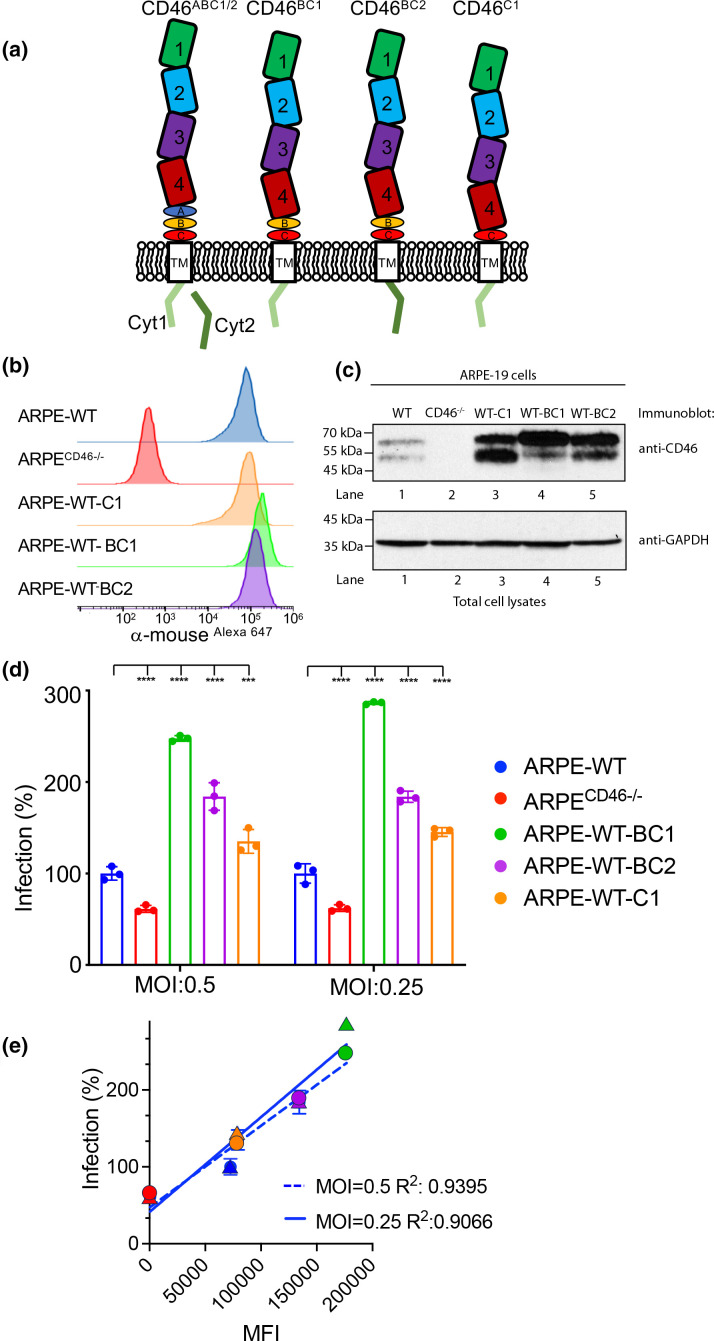
Fig. 2.
CD46 isomers enhance HCMV infection of ARPE-19 cells. (a) Schematic depicting the CD46 isomers. (b) Cell surface expression of CD46 in WT ARPE-19, ARPECD46-/- knockout cells, and WT ARPE-19 cells stably expressing CD46 C1 (ARPE-WT-C1), BC1 (ARPE-WT-BC1), and BC2 (ARPE-WT-BC2) was examined by fluorescence-based flow cytometry using anti-CD46 mAb GB24 followed by incubation with anti-mouse IgGAlexa647. The normalized cell number was plotted based on Alexa647 fluorescence intensity. (c) The cell lysates from the WT ARPE-19, ARPECD46-/-, ARPE-WT-C1, ARPE-WT-BC1, and ARPE-WT-BC2 cells were subjected to anti-CD46 and anti-GAPDH immunoblots, respectively. (d) WT ARPE-19, ARPECD46-/-, ARPE-WT-BC1, ARPE-WT-BC2, and ARPE-WT-C1 cells infected with AD169R (MOI:0.5 and 0.25) were assessed at 24hpi by a Celigo cytometer for infection. Percent infection was measured based on the anti-IE1 immunostain and WT ARPE-19 cells were used to normalize infection at 100 %. s.d. is depicted in the figure by errors bars. (e) A linear regression model was used to calculate the Goodness of Fit (R2 ) using infection (%) and mean fluorescence intensity (MFI) of CD46 surface expression of AD169R infection (MOI: 0.25 and 0.5). ***P<0.001, ****P<0.0001 (two-way ANOVA, Dunnett’s multiple comparison test) values are compared to ARPE-WT infection.