Figure 2.
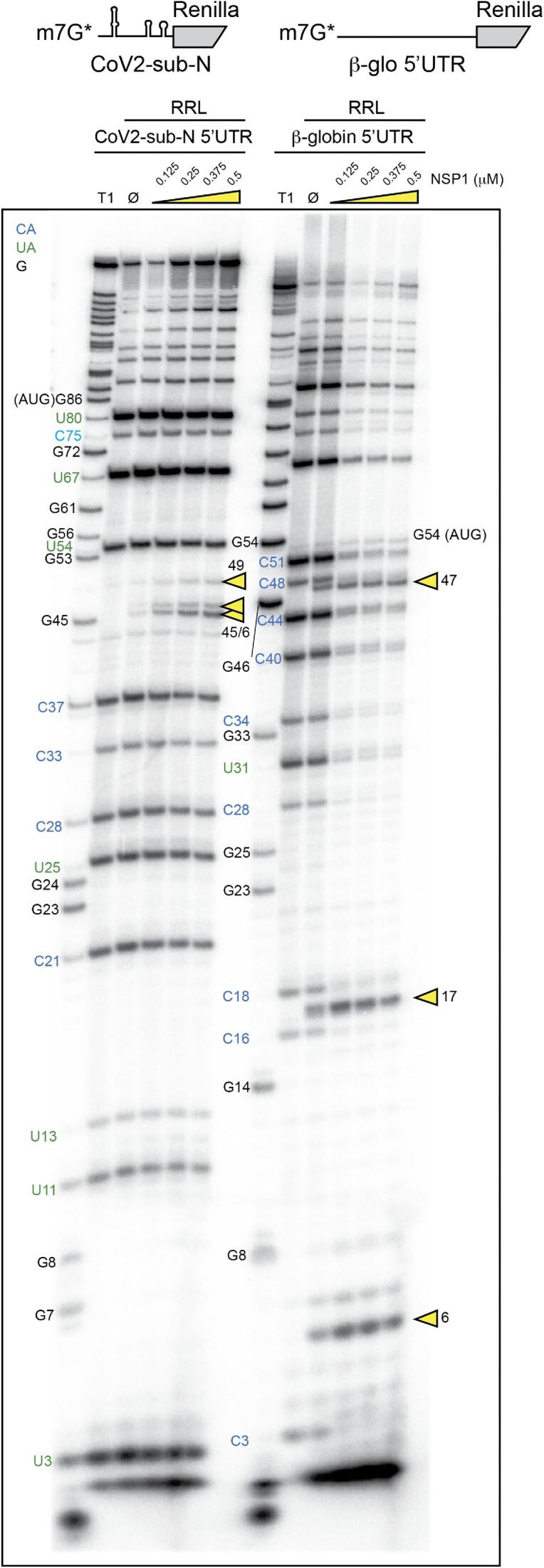
The cartoons represent the mRNAs used for the RNA degradation assay. SARS-CoV-2 subgenomic RNA (CoV2-sub-N) contains SL1–SL2–SL3 from the 5′UTR of the subgenomic RNA coding for the nucleocapsid N upstream of the N-terminal part of the Renilla luciferase coding sequence. The second RNA contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. Both RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs in RRLs for 5 min at 30°C in the absence (Ø) or presence of 0.125, 0.25, 0.375 and 0.5 μM wt SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. A T1 ladder made by RNase T1 digestion is loaded in parallel to map the cleavage site positions (T1). The cleavage sites are indicated by yellow arrowheads.
