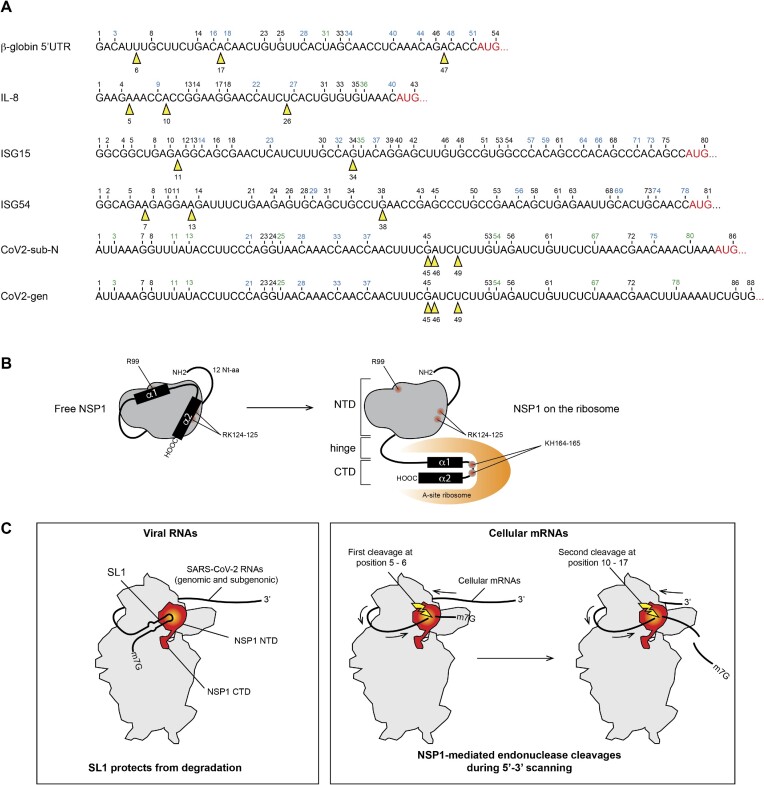
Figure 9.
(A) The sequences of the 5′UTR tested in this study are shown. The black numbers correspond to the position of G residues that are detected by RNase T1 digestion in the T1 ladder. The positions of NSP1-mediated cleavages are shown by yellow arrowheads. (B) Cartoon showing the NSP1 topology. In a free state, helices α1 and α2 are positioned on the N-terminal globular domain (grey) of NSP1, thereby masking residues R99, R124 and K125. NSP1 binding to the ribosome induces an important structural rearrangement; helices α1 and α2 interact specifically with the ribosome mRNA channel, which liberates the residues R99, R124 and K125 that are critical for endonuclease activity. (C) Model of endonuclease cleavage of NSP1 on the ribosome. NSP1 (in red) binds to the 40S ribosomal subunit (in grey). The NTD binds to the beak of the 40S subunit and the CTD binds in the mRNA channel and inhibits translation. The SL1 hairpin that is present on viral RNAs interacts directly or indirectly with NSP1 and thereby blocks the endonuclease cleavage site that is located in the NTD (in yellow). Cellular mRNAs do not contain SL1; they undergo endonucleolytic cleavages during 5′–3′ scanning. The first cleavage (shown by a yellow flash) occurs at position 5–6, then during subsequent sliding of the RNA, the second cleavage occurs at position 10–17.