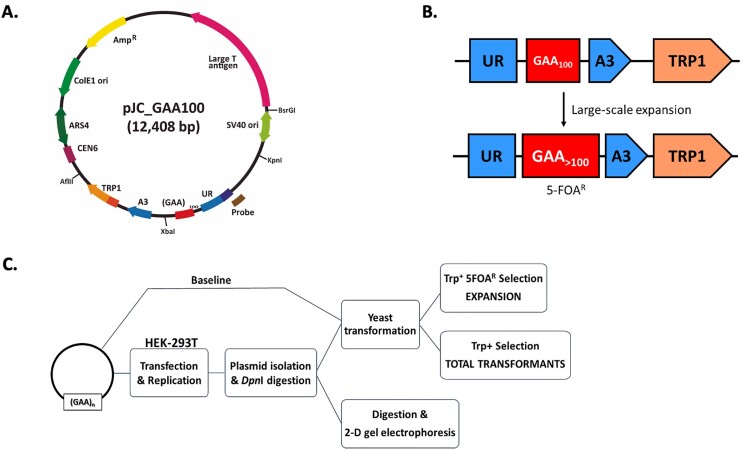
Figure 1.
An experimental system to study genome instability and fork stalling caused by GAA•TTC repeats. (A) pJC_GAA100 plasmid used in this study. The relative positions of its most relevant features are indicated inside: The centromeric sequence CEN6, the Autonomous Replication Sequence (ARS4), the ColE1 unidirectional origin (ColE1 Ori), the ampicillin resistance gene (AmpR), the Large T antigen gene, the SV40 origin of replication (SV40 ori) and the selectable cassette for repeat expansions (UR-GAA100-A3-TRP1: depicted in B). Outside, the relative positions of sites recognized by specific restriction endonucleases are indicated. (B) Schematic of the system to select for repeat expansion in yeast. An artificially split URA3 gene contains 100 GAA•TTC repeats such that expansion events abrogate splicing and result in resistance to a 5-fluoroorotic acid (5-FOA-r). The addition of more than 10 repeats increased the overall length of the intron beyond the splicing threshold. The selectable cassette was cloned into the pJC_GAA100 plasmid. TRP1 - an auxotrophic marker for the selection of yeast transformants bearing the plasmids. (C) Schematic representation of the assay. Plasmids for the study were transfected into HEK-293T cells, and after culturing them for 48 h, DNA was isolated and digested with DpnI. Repeat expansions were detected upon DNA transformation into yeast. Single-colony PCR was performed for 5-FOA-r clones to confirm expansion events. Expansion frequency was calculated by dividing the number of colonies with repeat expansions by the total number of TRP+ transformants. To calculate background expansion frequency, pJC_GAA100 plasmid isolated from E. coli was transformed directly into yeast. To study DNA replication through the repeats, DNA was digested with appropriate restriction enzymes and replication intermediates were analyzed by 2-dimensional (2D) agarose gel electrophoresis.