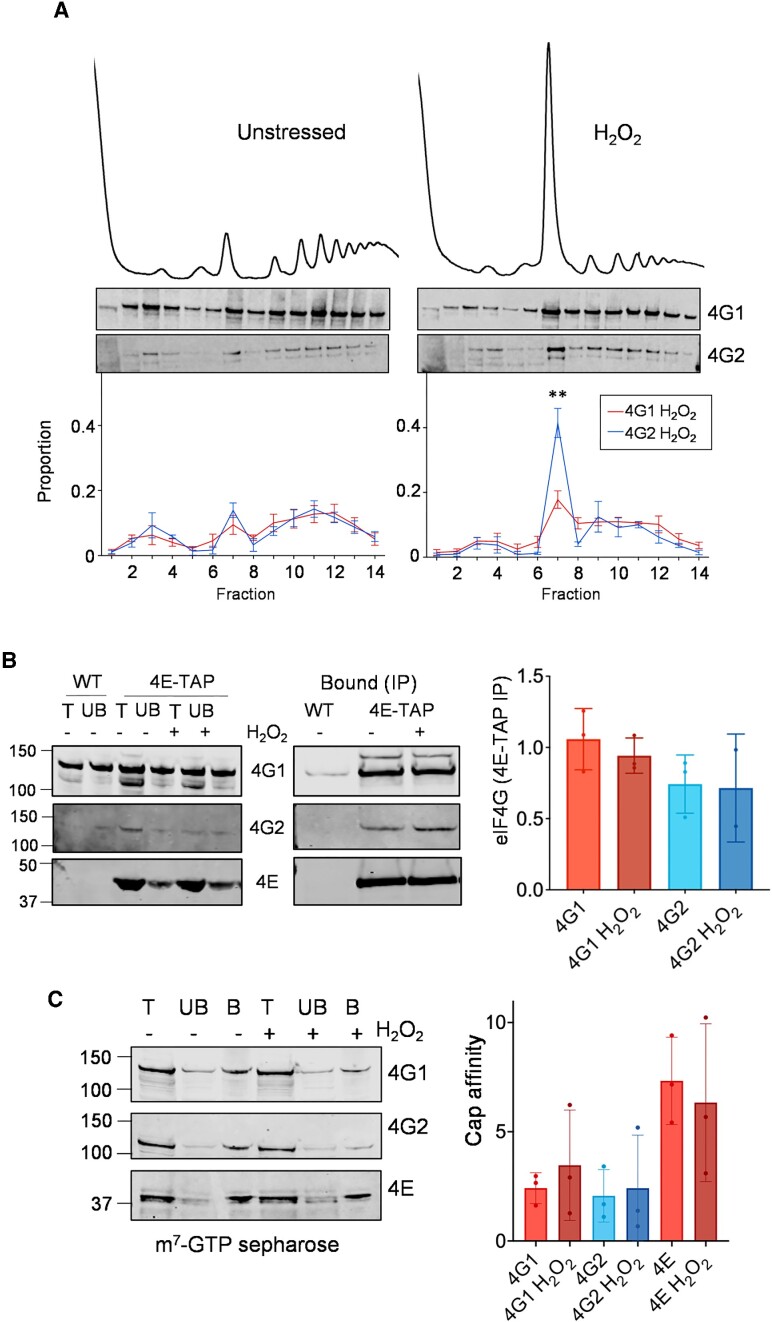
Figure 5.
eIF4G1 is associated with actively translating mRNAs following oxidative stress conditions. (A) Polyribosome profiles are shown for wild-type strain before or after hydrogen peroxide treatment for 15 min. Sucrose density gradients were fractionated and proteins visualised by Western blotting using antibodies specific for eIF4G1 or eIF4G2. The intensities of eIF4G1 and eIF4G2 bands were quantified and are shown as a percentage of the total cumulative intensity across each polyribosome trace. Quantification is shown from triplicate experiments (pairwise t-test, **P < 0.001) (B) eIF4E-TAP was immunoprecipitated from an exponential phase whole cell extract either unstressed or treated with 0.4 mM H2O2 for 15 min. The total (T), unbound (UB) and bound (IP) fractions were analysed by western blotting and membranes probed with antibodies for eIF4E, eIF4G1 and eIF4G2. Quantification is shown for the normalised ratio of eIF4G: eIF4E from triplicate experiments. No significant differences were found using one-way ANOVA. (C) A whole cell extracts were prepared from the wild-type strain either unstressed or treated with 0.4 mM H2O2 for 15 minutes and incubated with m7GTP-sepharose. Total (T), unbound (UB), and bound (B) fractions were analysed by Western blotting. Membranes were probed with antibodies for eIF4G1, eIF4G2 and eIF4E. Quantification is shown for bound factions as a percentage of totals. No significant differences were found using one-way ANOVA.