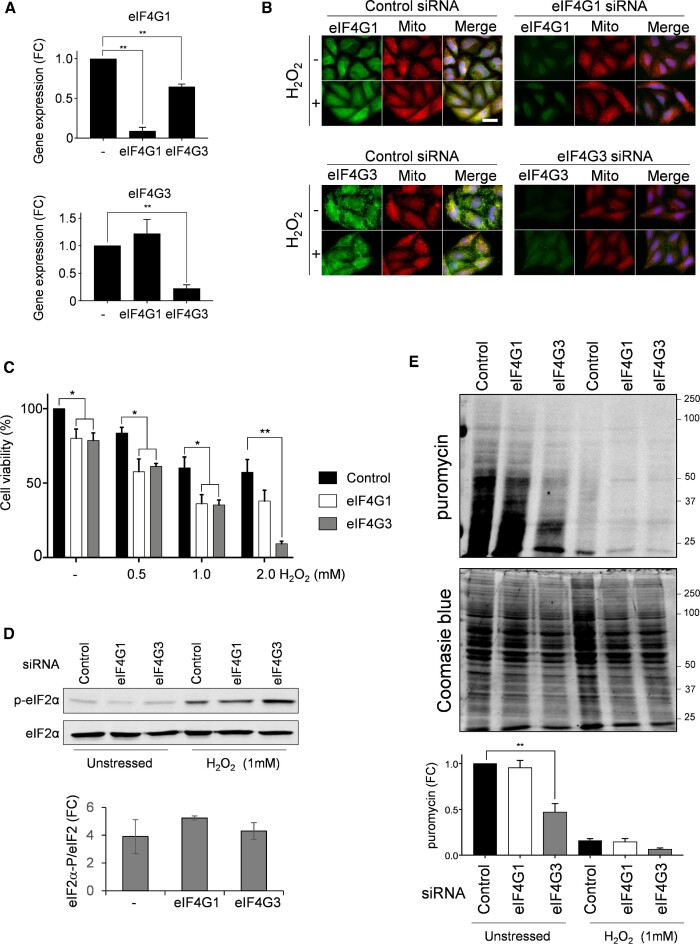
Figure 7.
Isoform specific requirements for eIF4G in human cells during oxidative stress conditions. (A) qRT-PCR analyses are shown to confirm the degree of specificity for eIF4G1 and eIF4G3 knockdowns. eIF4G3 siRNA specifically lowers eIF4G3 mRNA levels without affecting eIF4G1, whereas, eIF4G1 siRNA treatment strongly reduces eIF4G1 mRNA levels and decreases eIF4G3 expression by approximately 40%. Statistical analysis was carried out using one-way ANOVA. (B) The indicated HeLa cells were left untreated or treated with 1 mM hydrogen peroxide for one hour. eIF4G1 and eIF4G3 were visualized by immunofluorescence staining using specific antibodies (green). Mitochondria were visualized using MitoTracker Red and merged images are shown. (C) HeLa cells were treated with the indicated concentrations of hydrogen peroxide and viability measured using crystal violet. Statistical analysis was carried out using one-way ANOVA to compare the viability of the control and knock-down cells. (D) HeLa cells were left untreated or treated with 1 mM hydrogen peroxide for one hour. Protein extracts were immunoblotted for phosphorylated eIF2α (p-eIF2α) or eIF2α. Quantification is shown from triplicate experiments comparing the fold change (FC) increase in phosphorylation in response to hydrogen peroxide exposure. (E) Protein synthesis was analysed using puromycin labelling under the same conditions as shown in panel D. The incorporation of puromycin into newly synthesised protein was assessed by immunoblotting. Quantification is shown from triplicate experiments normalised against the intensity of Coomassie blue staining presented as fold change (FC) in puromycin labelling.