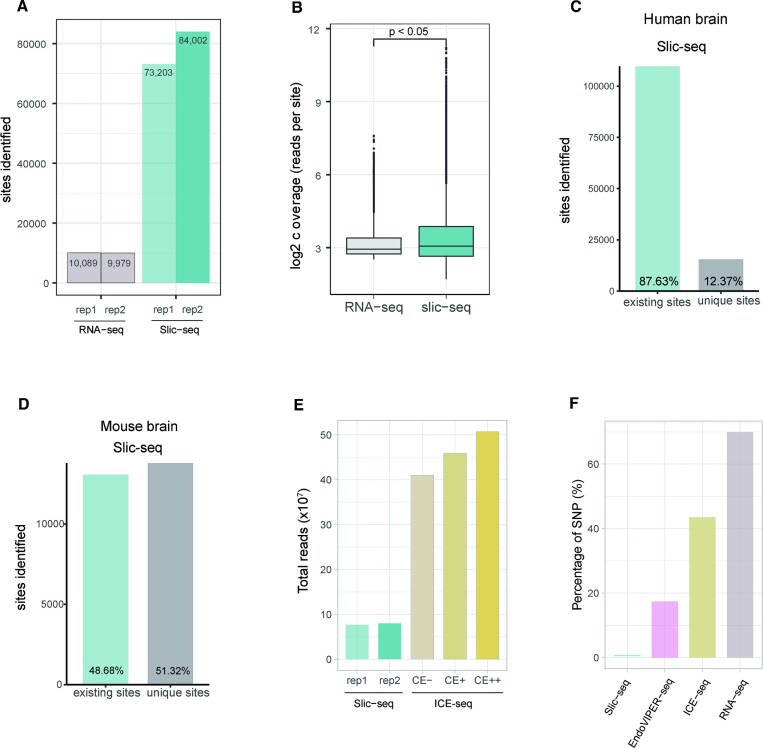
Figure 3.
Comparison of A-to-I editing sites identified by Slic-seq and other high-throughput sequencing methods. (A) Number of identified sites from RNA-seq and Slic-seq at the same sequencing depth; (B) Box and Whisker plot shows that read coverages at A-to-I editing sites are significantly increased by Slic-seq (two-sided t-tests, P < 0.05); (C) the proportion of A-to-I editing sites identified by Slic-seq from human brain in (existing sites) or not in (unique sites) REDIportal; (D) the proportion of A-to-I editing sites identified by Slic-seq from mouse brain in (existing sites) or not in (unique sites) REDIportal; (E) fractions of all sequenced read pairs (Slic-seq and ICE-seq) before trimming and mapping; (F) SNPs’ fractions of A-to-G candidate sites for HEK293T in RNA-seq, EndoVIPER-seq and Slic-seq.