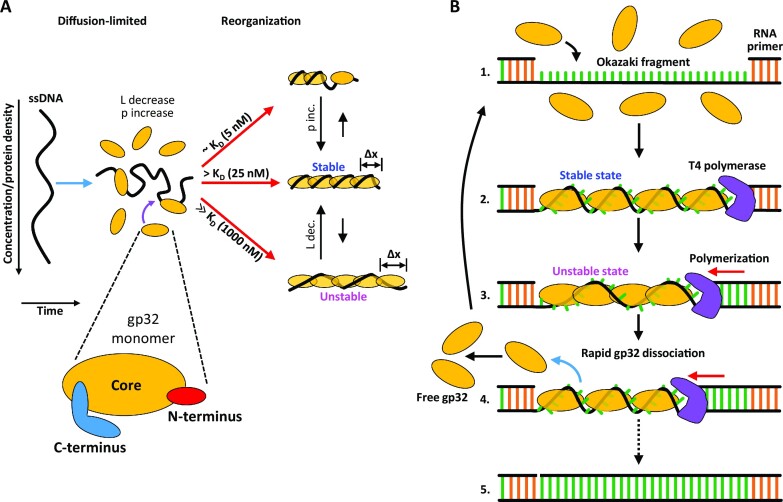
Figure 10.
gp32 binding states and function. (A) Diagram illustrating the different concentration-dependent gp32 binding states and pathways measured in this study. gp32 binding reduces the contour length (L) and increases the persistence length (p) of ssDNA. At gp32 concentrations approximately equal to KD (∼5 nM), gp32 filamentation along the ssDNA is incomplete. At [gp32] > KD (∼25 nM), the DNA is optimally saturated and filamented with gp32, giving rise to an increase in persistence length as the complex reorganizes into its most stable conformation. At protein concentrations well above saturating (∼1000 nM), the protein density along the DNA increases further, resulting in an increase in the protein-DNA contour length as the complex equilibrates to a more extended (Δx) and less stable conformation. (B) Diagram illustrating a model for the function of gp32’s unstable binding mode during DNA replication. During lagging strand synthesis, Okazaki fragments are formed (1) and subsequently coated with gp32 in a stable binding conformation (2). Polymerization along the strand drives an increase in protein density as the ssDNA segment shortens, forcing the gp32 filament to adopt a less stable conformation (3) that results in rapid protein dissociation and recycling (4). This process continues until the lagging strand is completely synthesized (5).