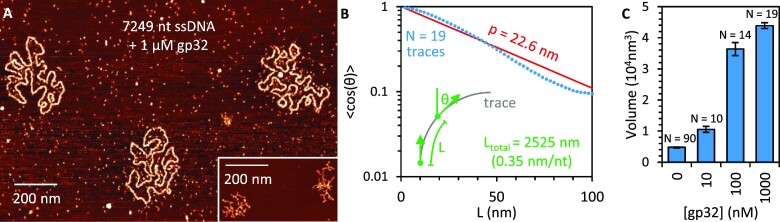
Figure 4.
AFM imaging of gp32–ssDNA complex. (A) AFM image of 7249 nt long ssDNA incubated with 1 μM gp32. While protein-free ssDNA (inset, same scale) is condensed due to its tendency to fold back on itself, a result of its short persistence length and the formation of secondary structure formed between complementary bases in different regions of the ssDNA, the protein-saturated ssDNA forms one long continuous filament that can be traced along the 2D surface. (B) Traces of individual molecules are used to measure the average value of the cosine of the change in orientation angle (θ) between any two points separated by a length (L) along the trace. Average cos(θ) decreases exponentially as L increases, consistent with the WLC model. Fitting this decay parameter yields an effective persistence length (red line). (C) The total integrated volume of ssDNA molecules incubated with varying concentrations of gp32 is measured as a proxy for total protein bound to the substrate. At high concentration, the volume increases nearly 10× as compared to protein-free ssDNA, indicating the ssDNA–gp32 complex is protein-saturated.