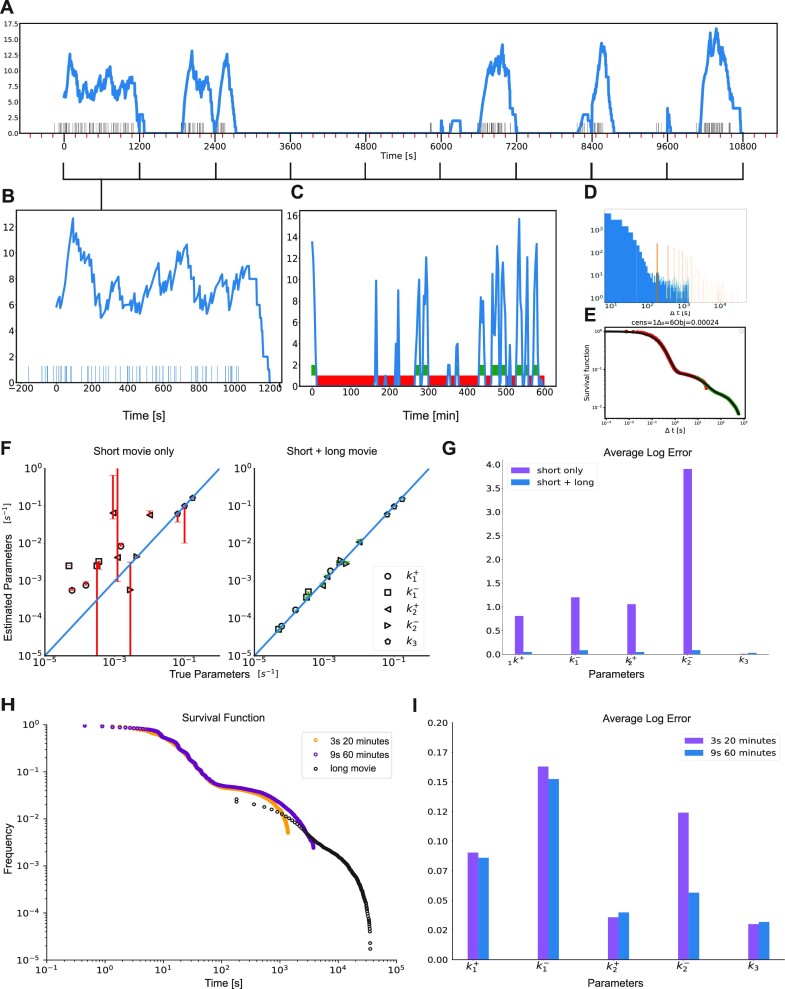
Figure 4.
Combining short and long movies. (A) Simulated long duration signal using the three-state model M2 and high temporal resolution, parameters corresponding to dataset D14. The timeline with black markers represent polymerase start time positions. The transcription site signal is represented in blue, using short movie high temporal resolution. The x-axis major tick marks in black (every 1200 seconds) represent the duration of a high resolution short movie (1 stack every 3 s for 20 min). The minor ticks in red represent 3 min marks, i.e. the resolution of a long duration movie (1 stack every 3 min). (B) Simulated low resolution short movie using the model M2. Blue bars represent polymerase positions found by GA after deconvolution (blow-up start of the signal from A). (C) Low resolution long movie with thresholding to extract off periods or waiting times. (D) Histogram of length of waiting times obtained from short movies (in blue) and long movies (in orange). (E) Matched Survival function of the long (green) and short movie (red) with overlap in the middle. (F) Accuracy of parameter reconstruction of the model M2 for the parameter sets D12–14 in Table 1 using only a short movie and a short + long movie. (G) Average logarithmic error of the parameter reconstruction of the model M2 for the parameter sets D12–14 in Table 1 using a short movie (20 min every 3 s) combined or not with a long movie (10 h every 3 min). (H) Long and short movie survival functions and their overlap for different lengths and resolutions of the short movie. (I) Average logarithmic error of the parameter reconstruction of the model M2 for the parameter sets D12–14 in Table 1 for different durations and resolutions of the short movie.