Table 1.
FSMM parameters used to generate the artificial datasets. Furthermore, the MS2 sequence and elongation rate parameters were nseq = 1292 bp (24×MS2), npost = 4526 bp, Vpol = 45 bp × s−1 for D1–5 and D7–9,nseq = 5800 bp (128×MS2), npost = 8300 bp, Vpol = 67 bp × s−1 for D6 and D10–14. D1–5 and D7–9 parameters come from the study of Drosophila promoters in (20). D12–14 is based on estimates of HIV-1 transcription bursting in human cells studied in (19). D6 and D10–11 come from estimates of bursting from wild-type and mutated EEF1A promoters inserted in human cell lines
| Dataset/Ref. | Parameters | ||||
|---|---|---|---|---|---|
| 2 states | k +[s−1] | k −[s−1] | k ini [s−1] | ||
| D1 (20) | 0.02036 | 0.00150 | 0.11432 | ||
| D2 (20) | 0.01117 | 0.00593 | 0.07637 | ||
| D3 (20) | 0.01189 | 0.01430 | 0.07745 | ||
| D4 (20) | 0.02439 | 0.00144 | 0.13397 | ||
| D5 (20) | 0.04169 | 0.00414 | 0.11277 | ||
| D6 | 0.00484 | 0.00025 | 0.113 | ||
| 3 states M2 |
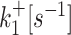
|
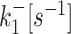
|
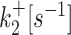
|
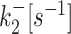
|
k ini [s−1] |
| D7 (20) | 0.01426 | 0.00339 | 0.06553 | 0.05751 | 0.17102 |
| D8 (20) | 0.00661 | 0.00013 | 0.05772 | 0.01054 | 0.13201 |
| D9 (20) | 0.00332 | 5.3 × 10−5 | 0.05804 | 0.00586 | 0.13119 |
| D10 | 0.0001 | 2.3 × 10−5 | 0.00091 | 0.00024 | 0.019 |
| D11 | 0.00023 | 3.2 × 10−5 | 0.0011 | 0.00019 | 0.018 |
| D12 (19) | 0.0015 | 4.9 × 10−5 | 0.01 | 0.0043 | 0.17 |
| D13 (19) | 0.00015 | 0.00031 | 0.0012 | 0.0028 | 0.1 |
| D14 (19) | 6 × 10−5 | 0.00035 | 0.00089 | 0.003 | 0.063 |
