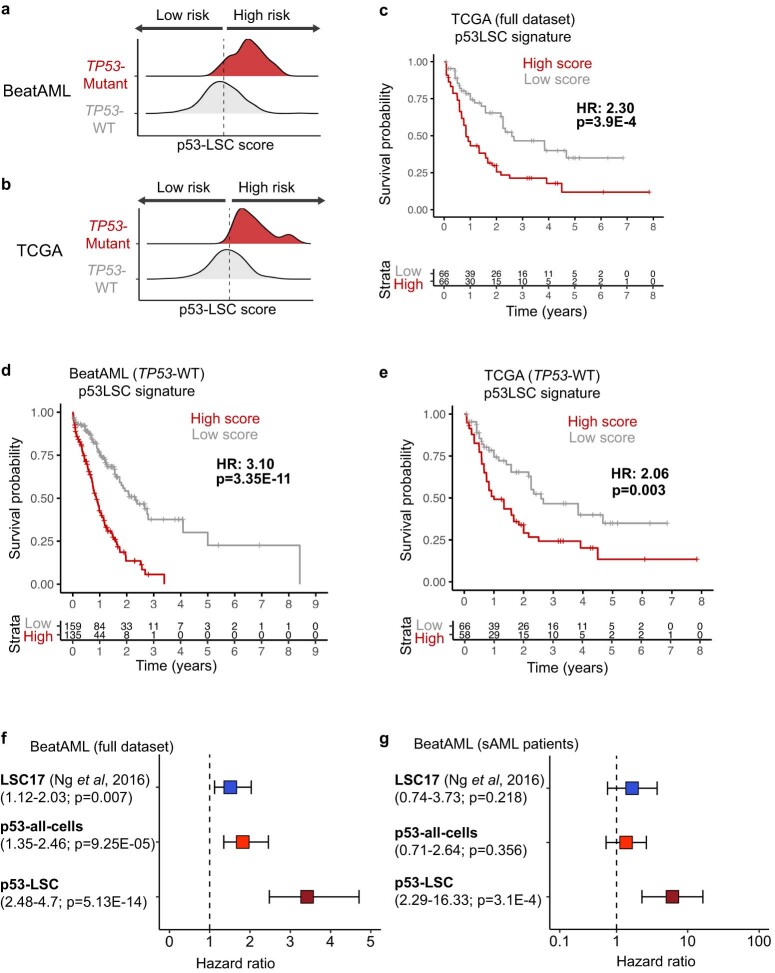
Extended Data Fig. 6. Validation of p53-LSC signature score in two independent cohorts.
a-b, Distribution of p53-LSC scores in BeatAML (a) and TCGA (b) cohorts stratified by TP53 mutational status. c-e, Kaplan-Meier analysis of de novo AML patients from the full TCGA AML dataset (n = 132) (c), TP53-WT AML patients from BeatAML (n = 294) (d) and TP53-WT AML patients from TCGA (n = 124) (e) stratified according to high or low p53 LSC signature score. f-g, Hazard ratio of all AML patients (n = 322) (f) or secondary AML patients (n = 49) (g) from the BeatAML cohort using LSC17 score (Ng et al, 2016), p53-all-cells score (derived from all TP53-mutant sAML cells) and p53-LSC signature score (derived from transcriptionally-defined LSCs; related to Fig. 2a). Boxes represent hazard ratios and lower and upper bounds of error bars, 95% confidence intervals. Genes used for each score are listed in Supplementary Table 4. “p” indicates log-rank test p-value.