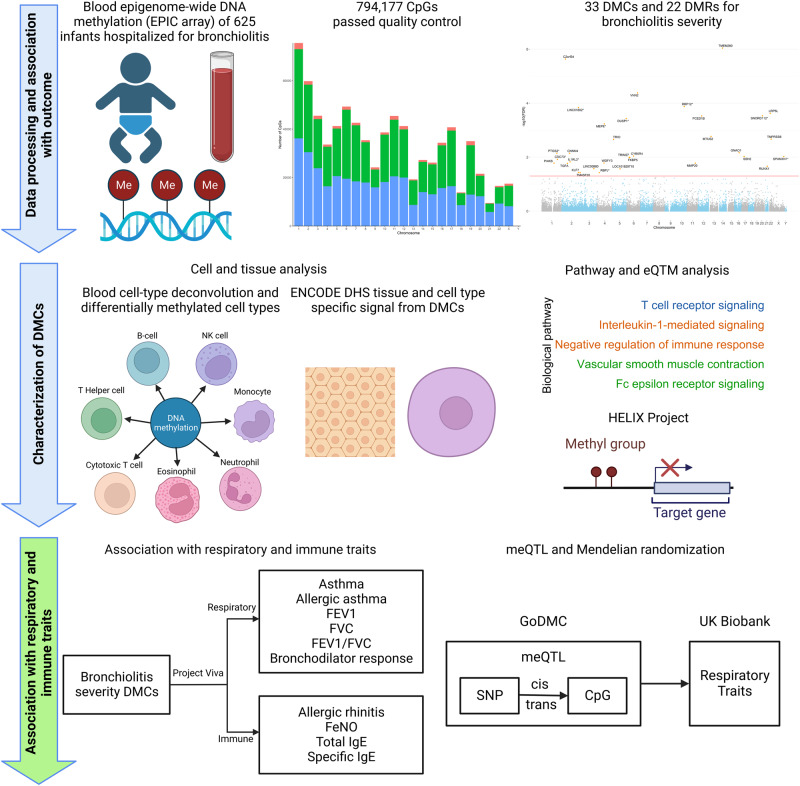
Fig. 1. Study Design and Analytic Workflow.
The analytical cohort consists of 625 infants hospitalized for bronchiolitis in a multicenter prospective cohort study—the 35th Multicenter Airway Research Collaboration (MARC-35). Blood Infinium MethylationEPIC array (850 K) DNA methylation data underwent quality control, leading to a total of 794,177 high-quality CpGs for the downstream analysis. In Aim 1, the association of 794,177 CpGs with the risk of PPV use was examined. A total of 33 severity-related DMCs and 22 DMRs were identified. In Aim 2, seven blood immune cell types were deconvoluted using the epigenome-wide DNA methylation data. The association of the DMCs with each cell type was examined. The ENCODE DHS tissue- and cell-specific signal from the DMCs was also determined. The biological pathway analysis using the GO, KEGG, and Reactome databases was performed. The association of blood DNA methylation and gene expression was investigated by cis-eQTM (HELIX Project) analysis. In Aim 3, by leveraging independent and publicly available EWAS (Project Viva) and GWAS (GoDMC and UK Biobank) data, the association of bronchiolitis severity-related DMCs with respiratory and immune traits was examined. Some components of this figure were created with BioRender.com. CpG, cytosine-phosphate-guanine; DHS, DNase hypersensitivity site; DMC, differentially methylated CpG; DMR, differentially methylated region; ENCODE, Encyclopedia of DNA Elements; eQTM, expression quantitative trait methylation; EWAS, epigenome-wide association study; GO, Gene Ontology; GoDMC, Genetics of DNA Methylation Consortium; GWAS, genome-wide association study; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPV, positive pressure ventilation.