Figure 1: Deep learning areas of focus in vaccine design.
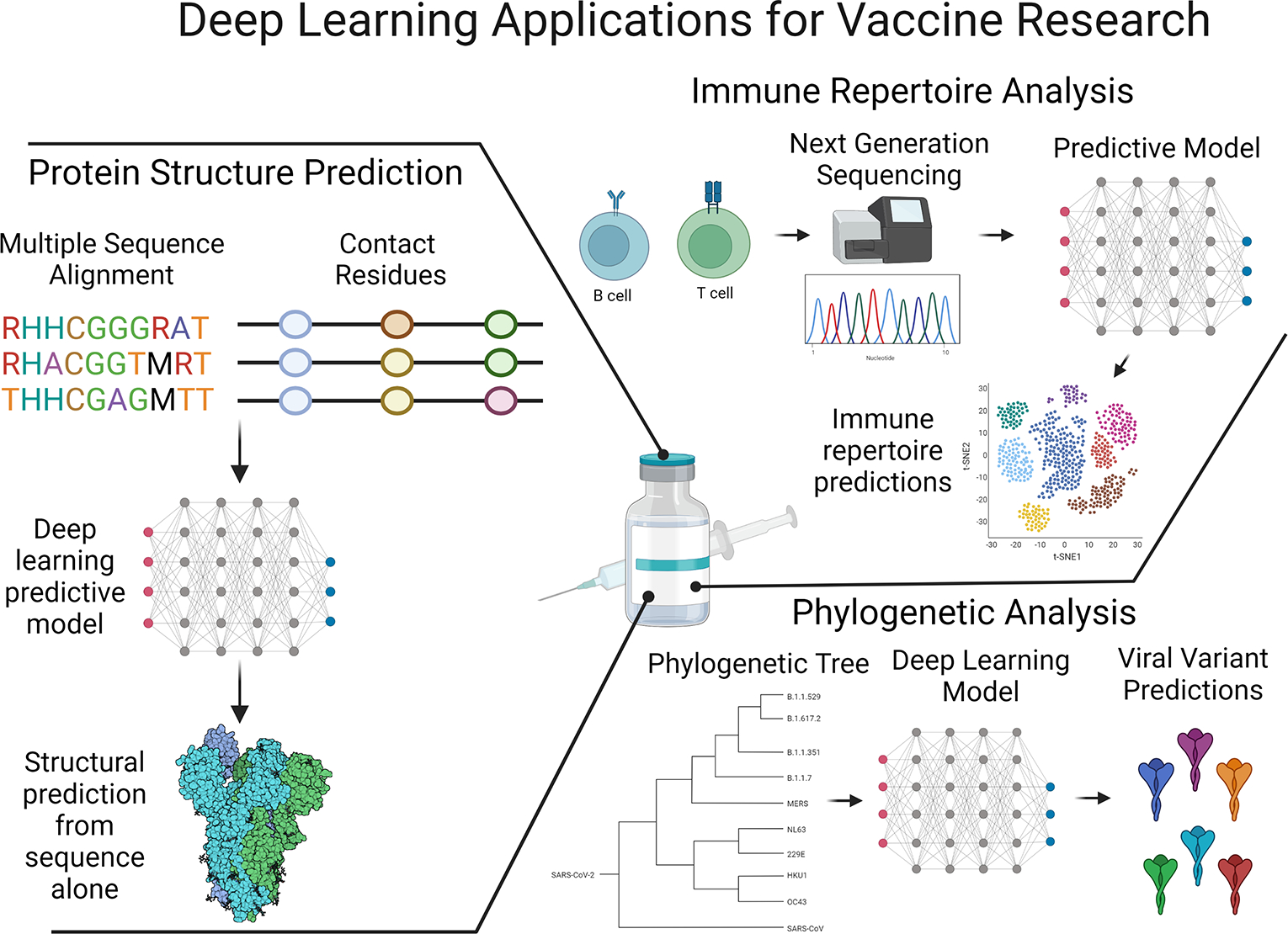
Prediction of protein structures, analysis of antibody and T cell receptor repertoires, and viral phylogenetics are three areas in which deep learning is supporting rapid advances. Deep learning has made the greatest progress so far in structure prediction “solving” the protein folding problem and is now commonly being used to generate antibodies bypassing experimentation steps. Immune repertoire data growth has coincided with deep learning development allowing for prediction of the specificity or disease outcomes of immune responses from sequencing data alone. Phylogenetic analysis of global viral variants can leverage deep learning to better understand mutational patterns and the effect mutations may have on subsequent immune responses as well as pathogen fitness and population susceptibility. This figure was created using BioRender (https://biorender.com/)
