Abstract
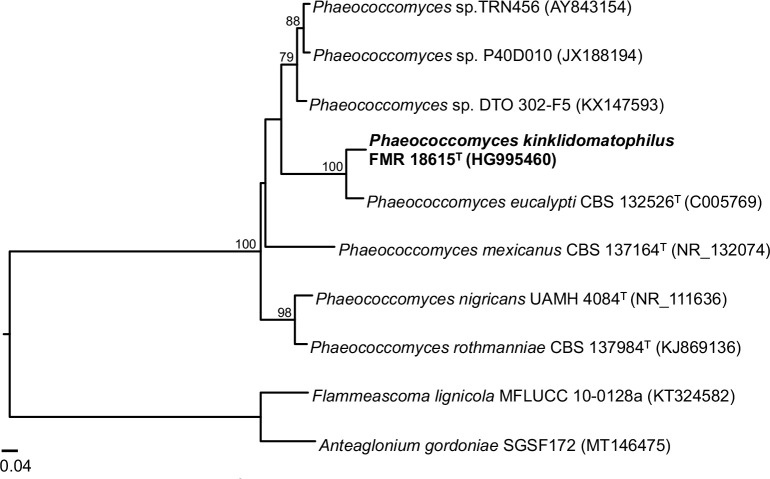
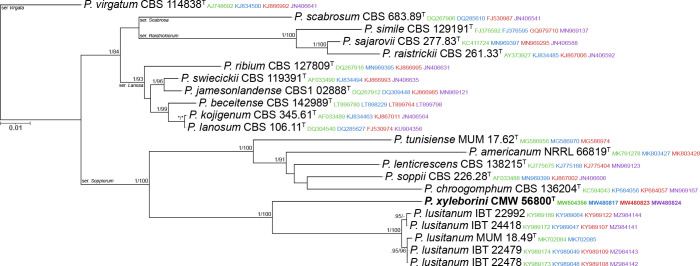
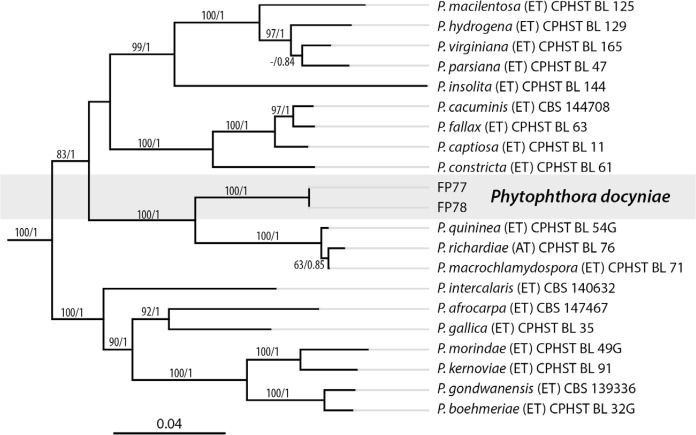
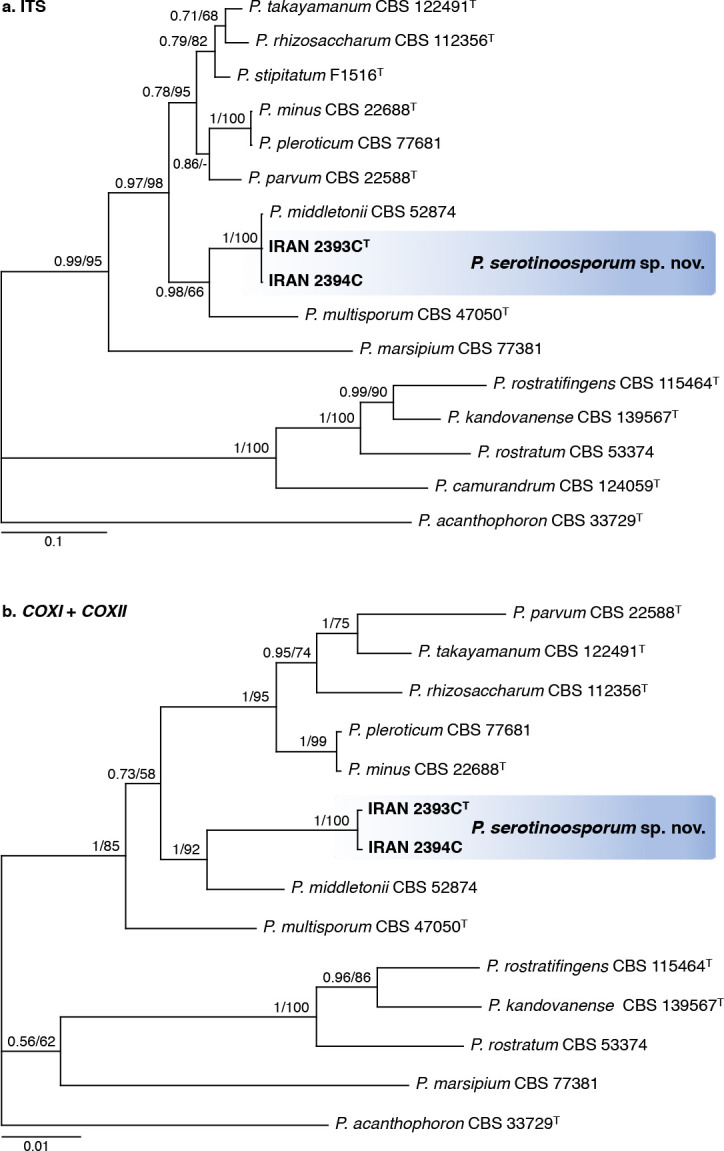
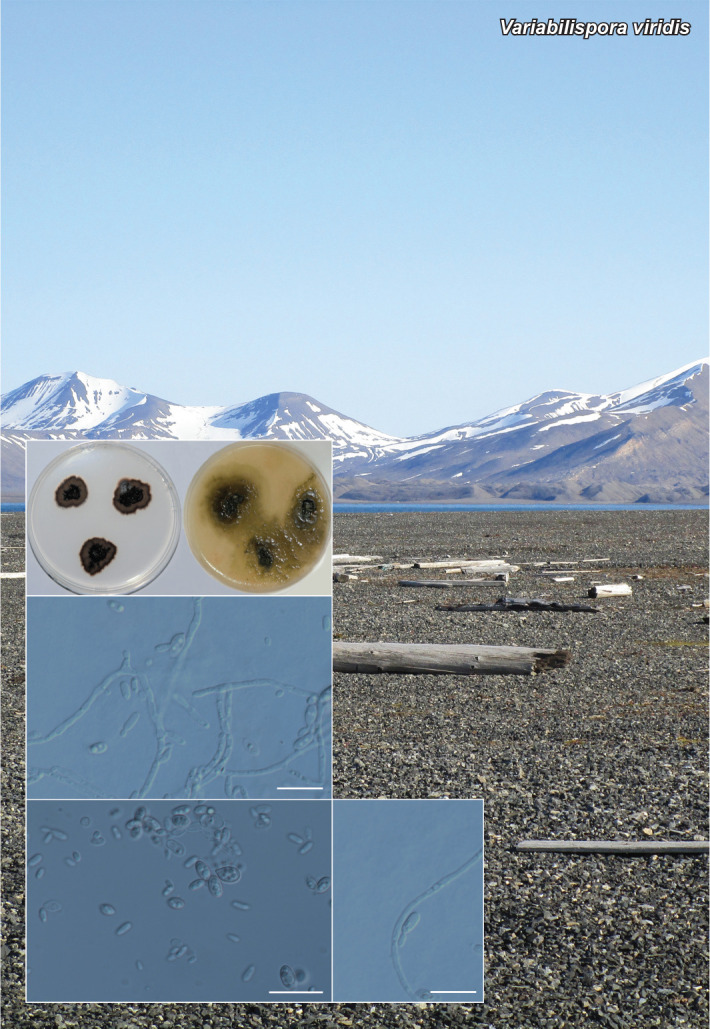
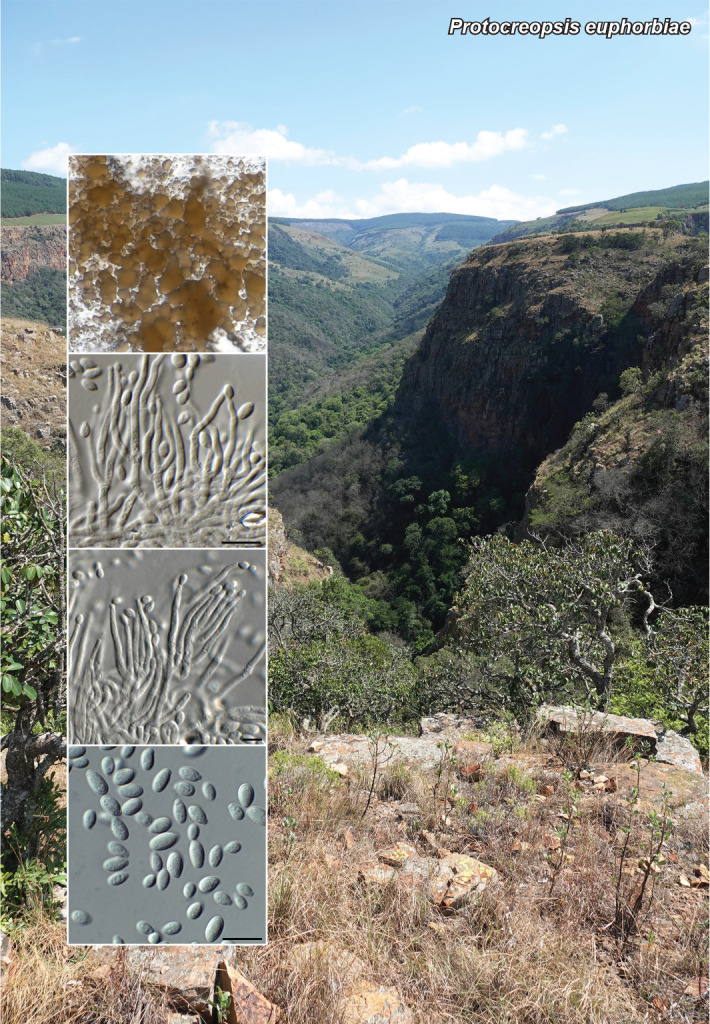
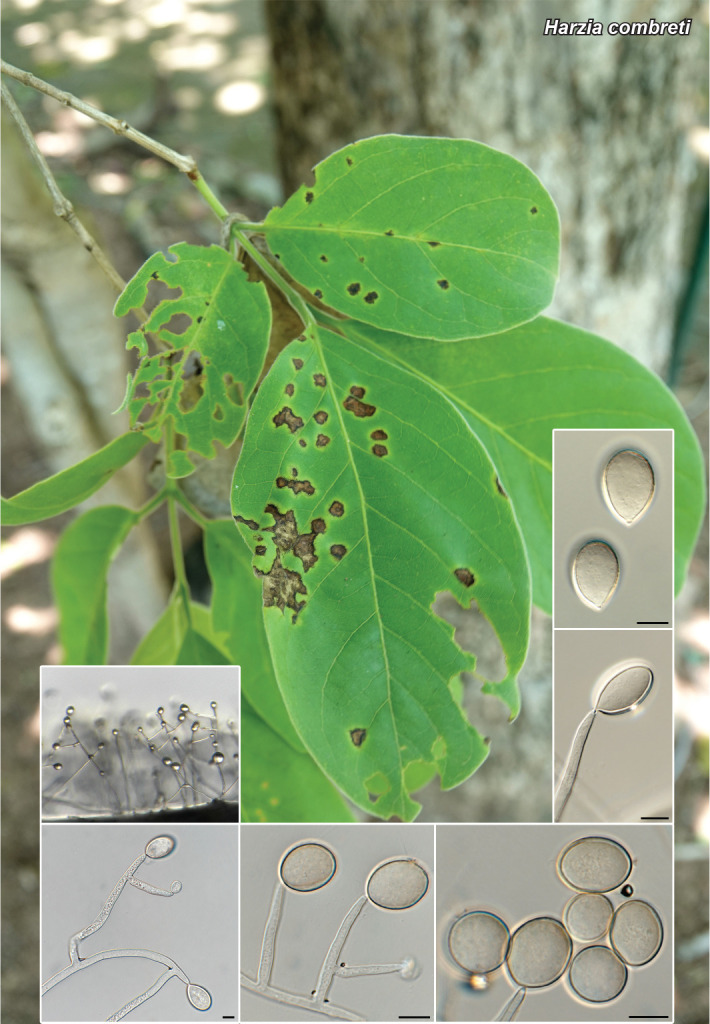
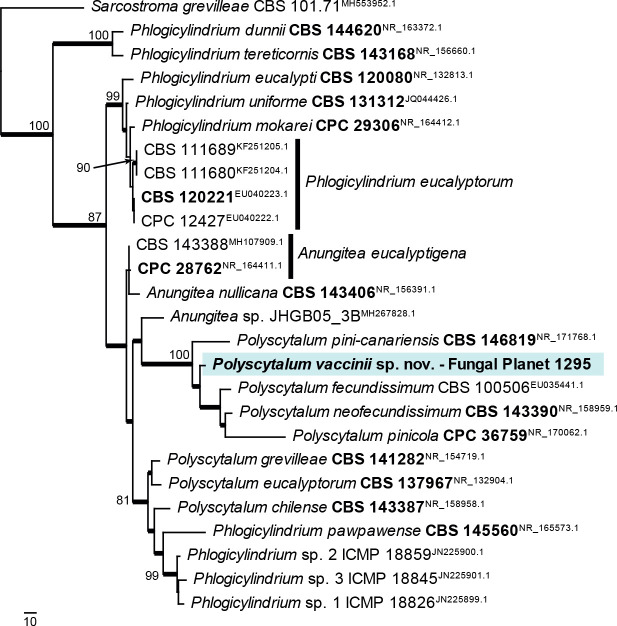
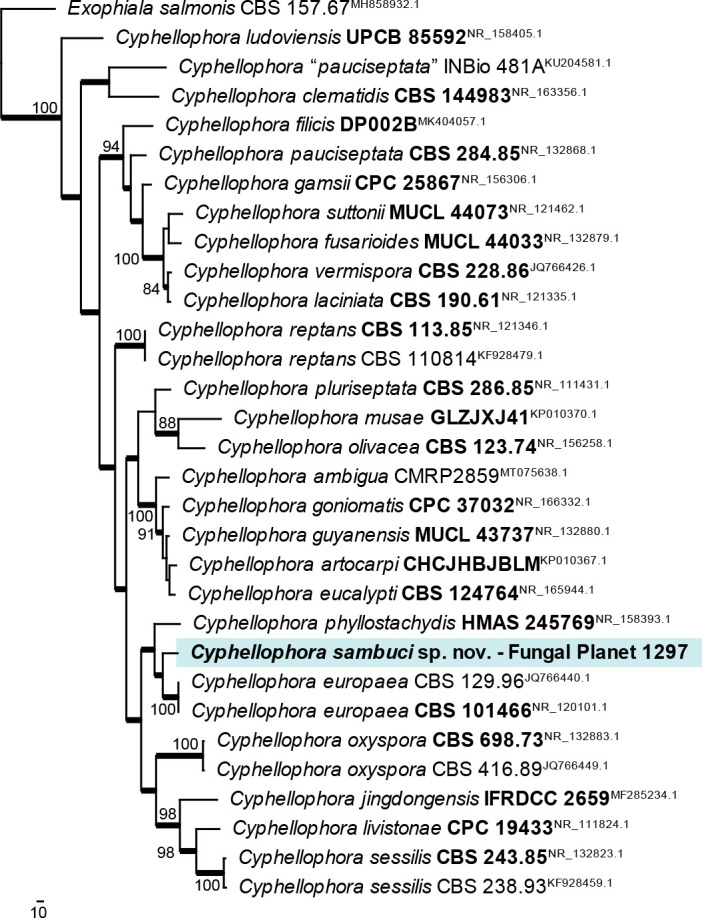
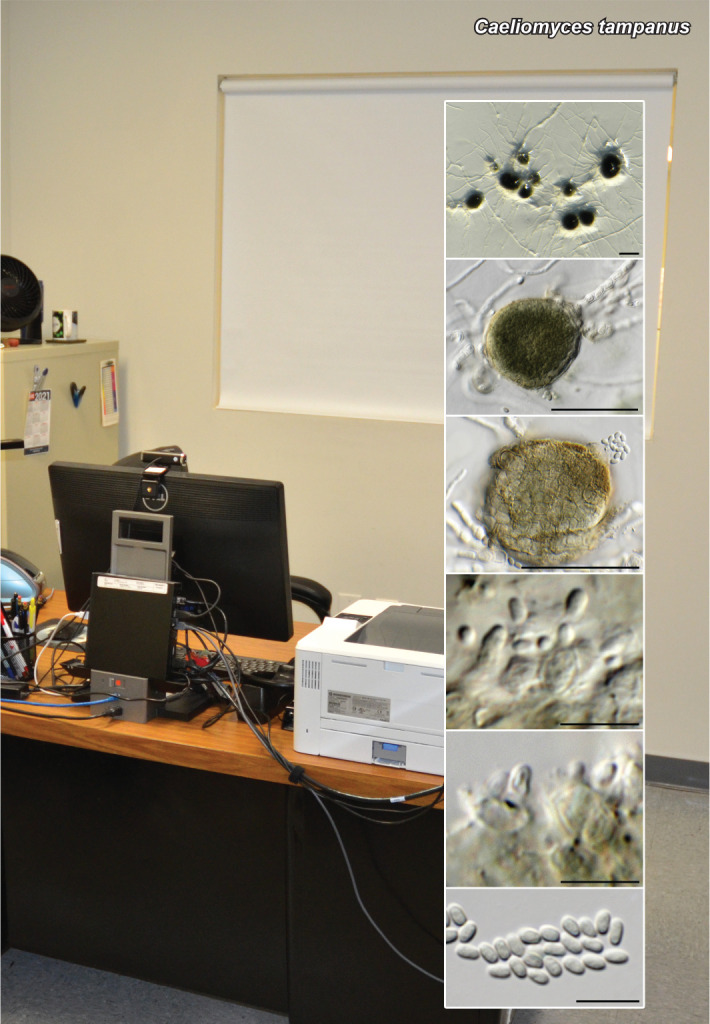
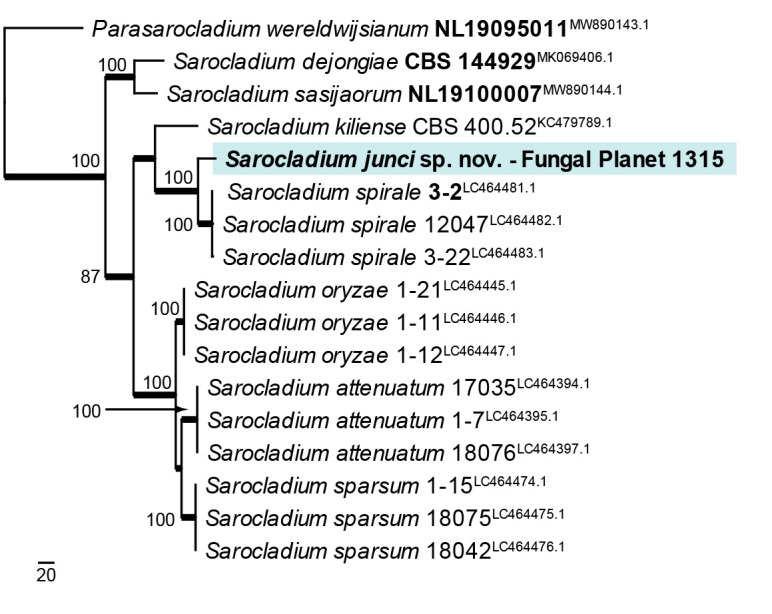
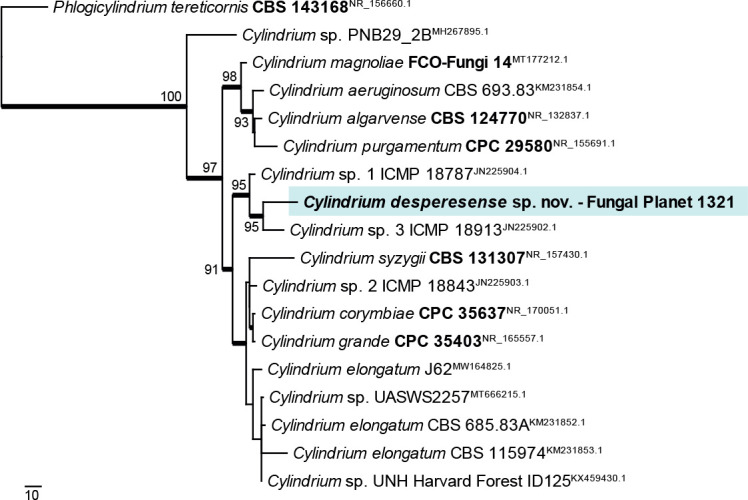
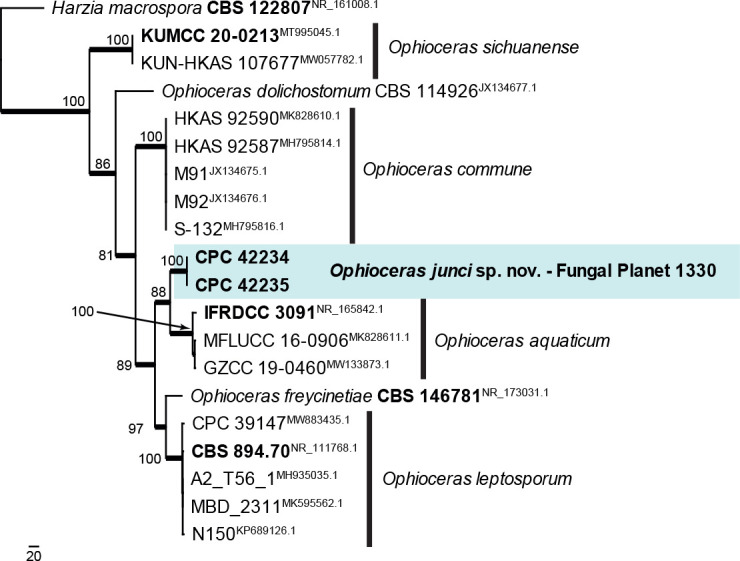
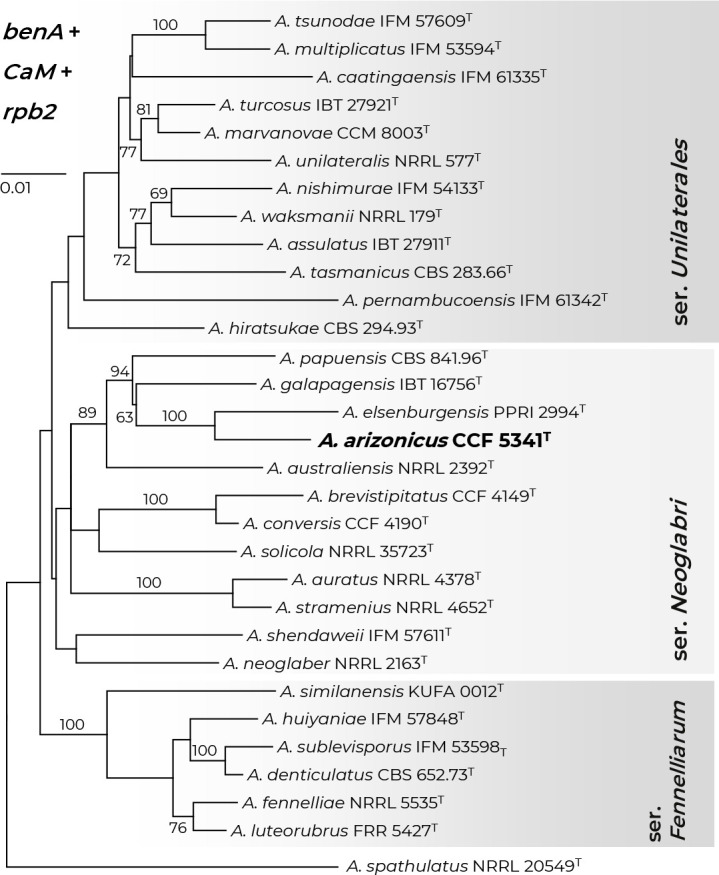
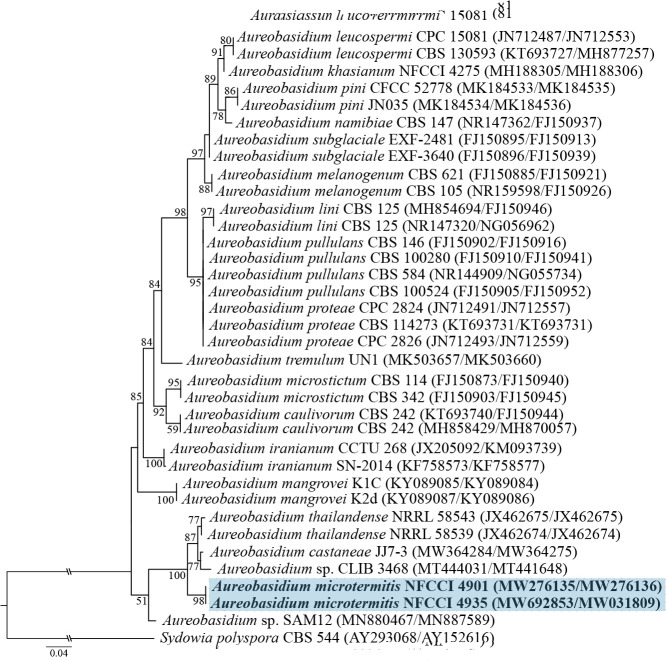
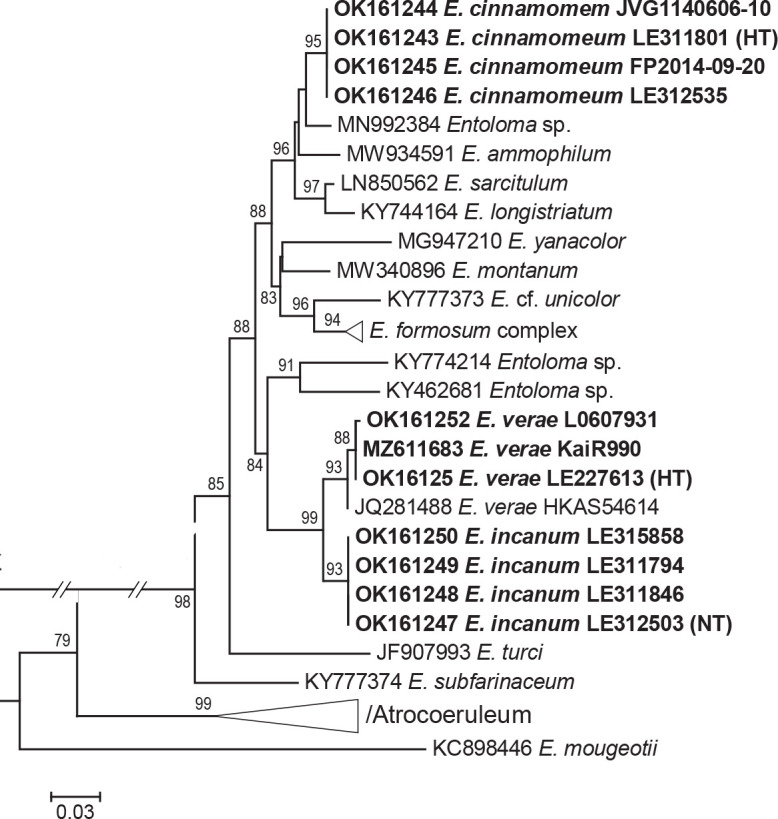
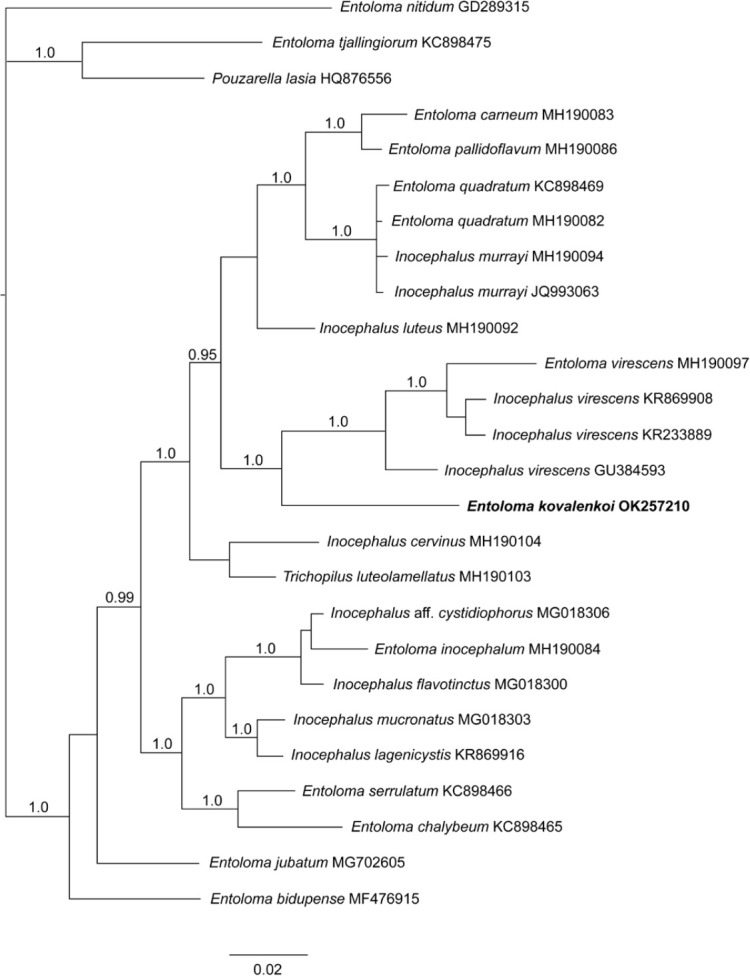
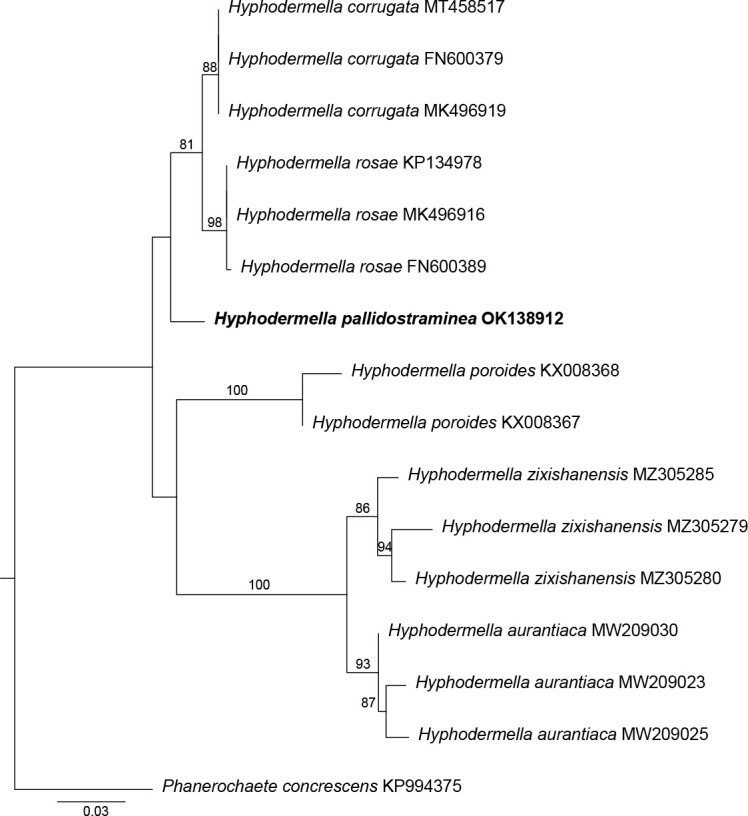
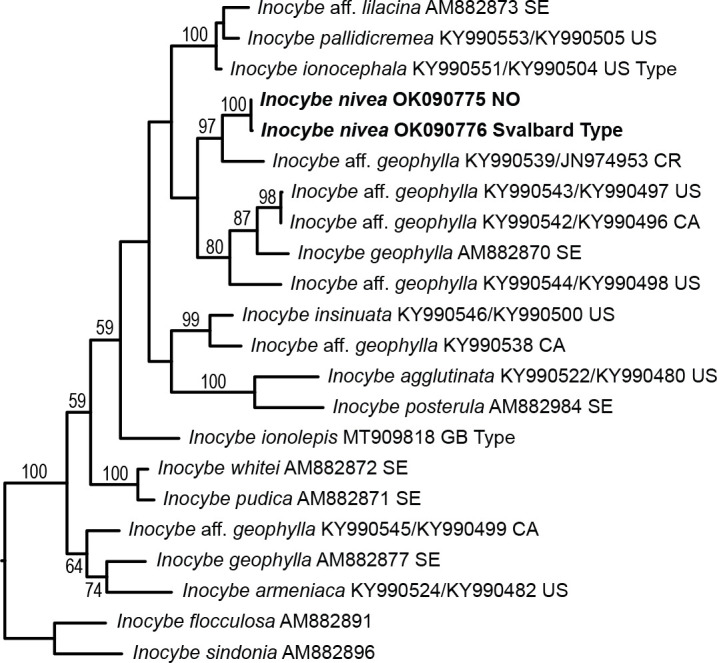
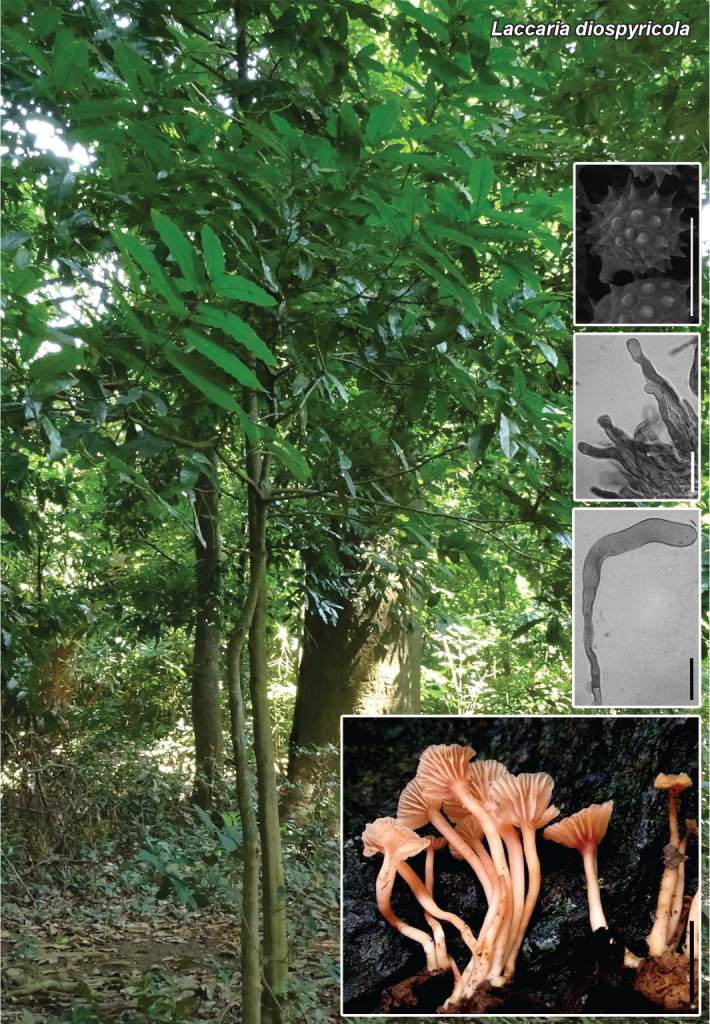
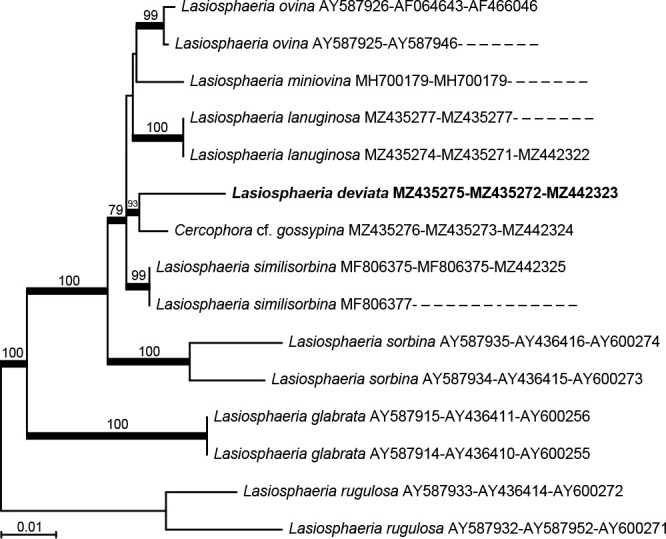
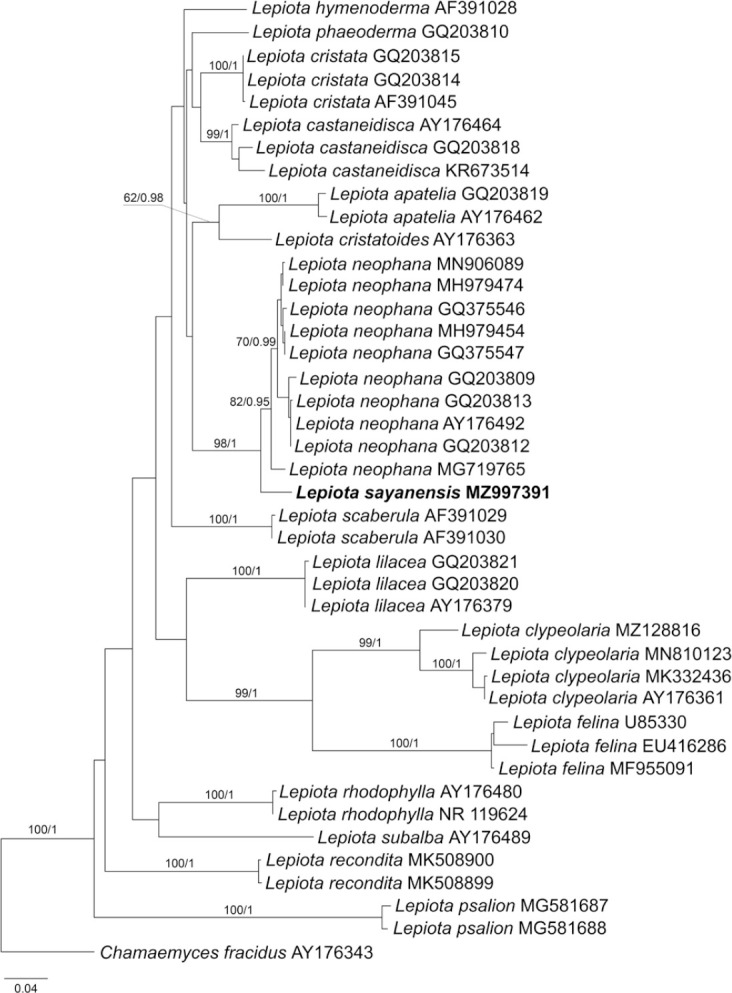
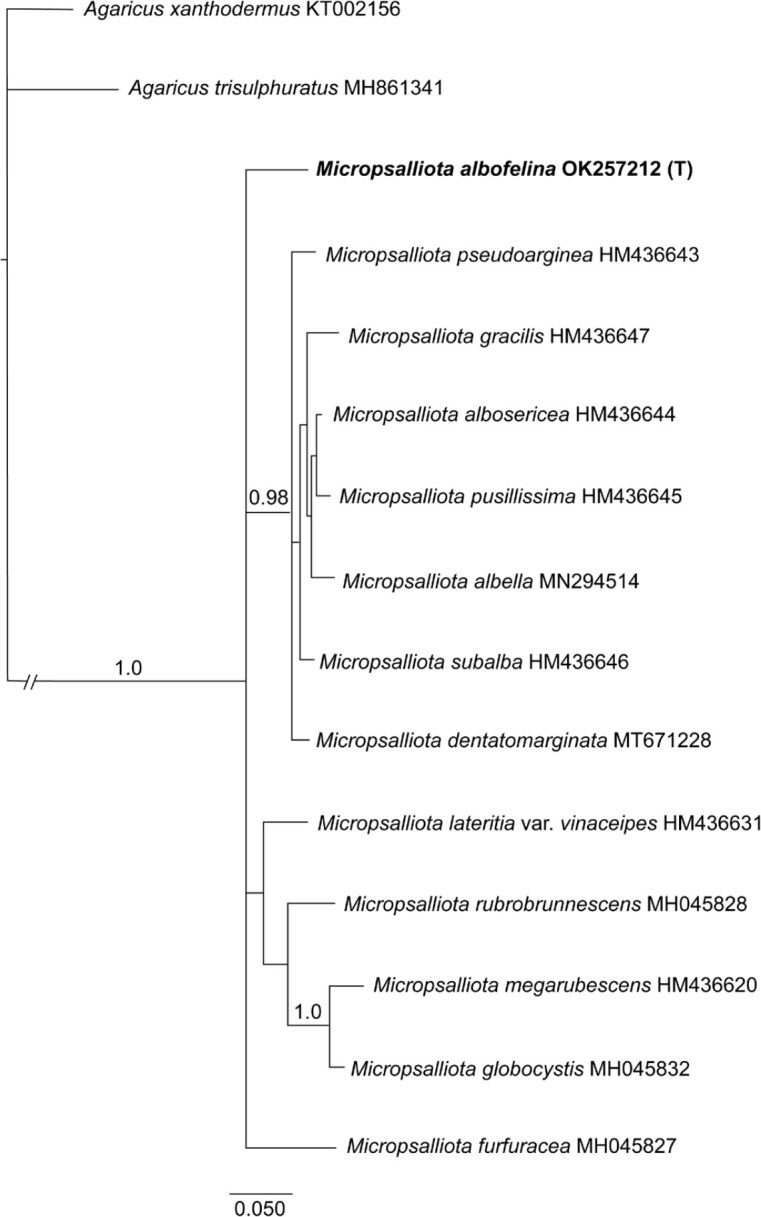
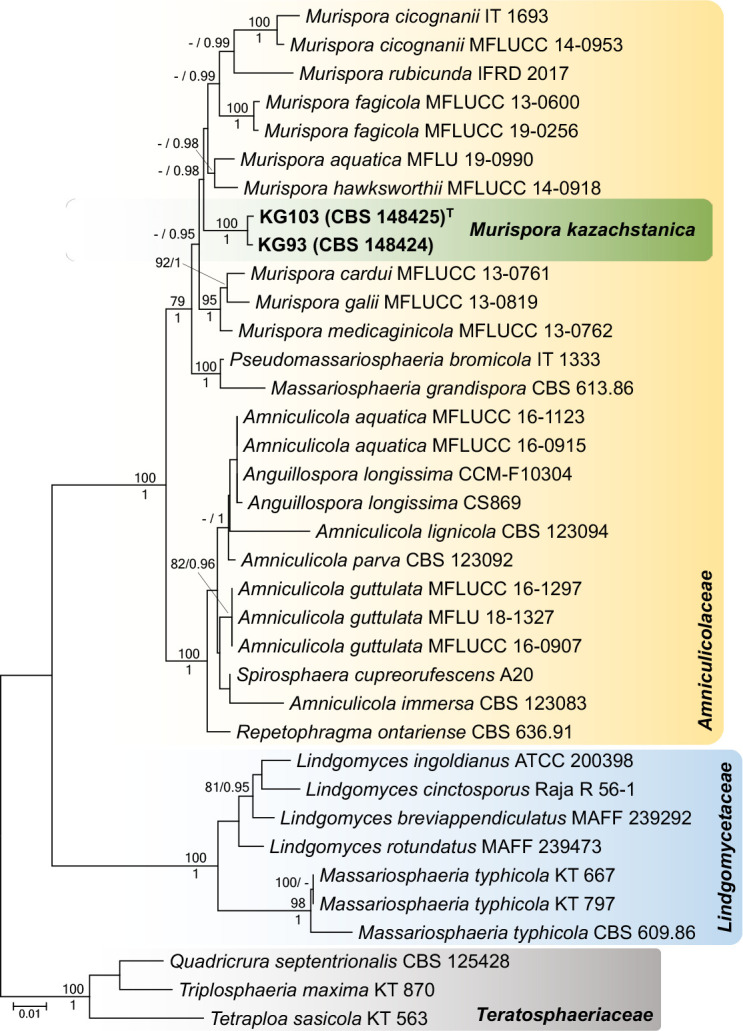
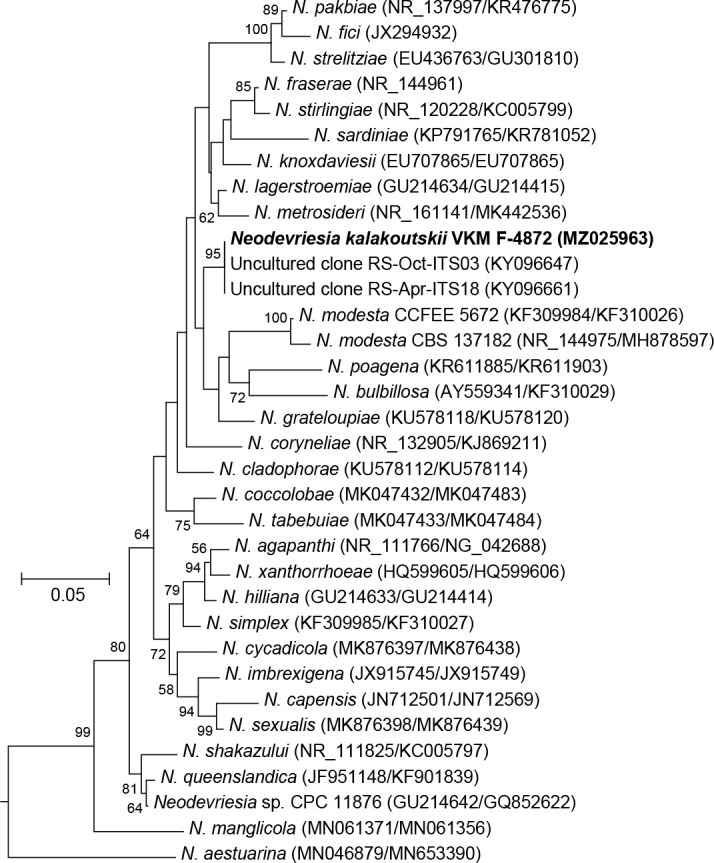
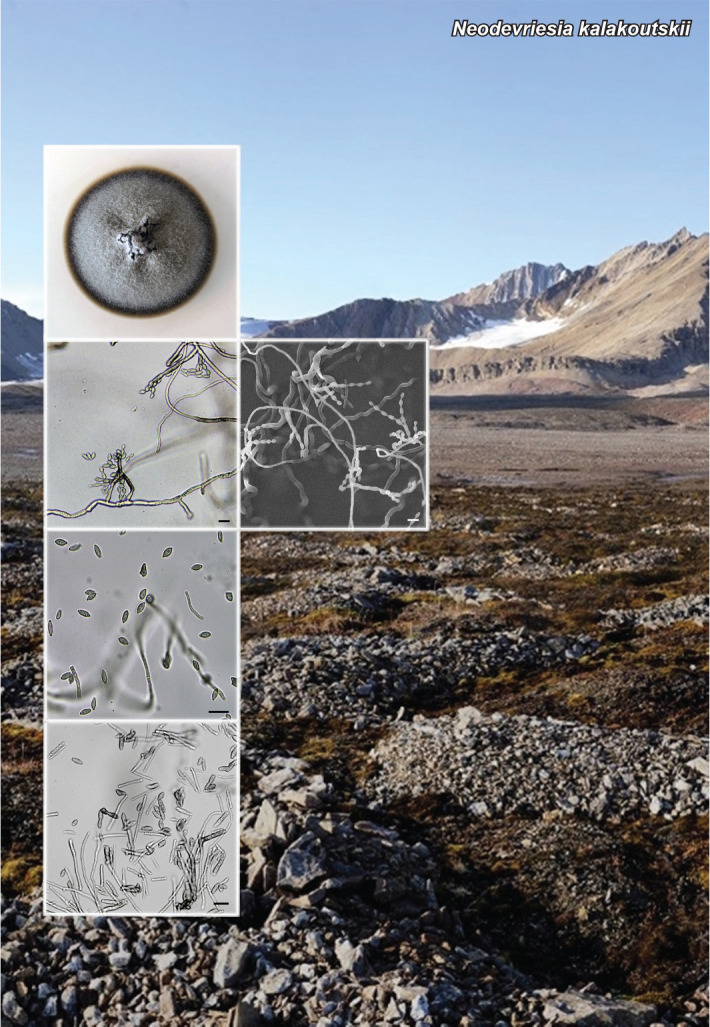
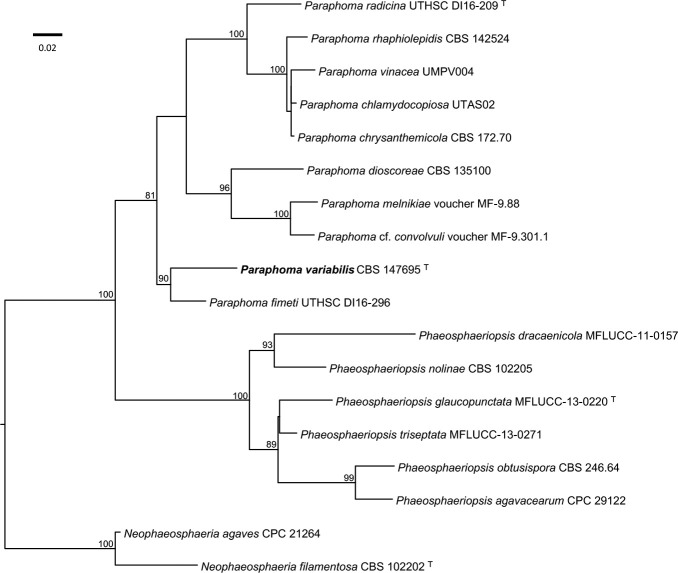
Novel species of fungi described in this study include those from various countries as follows: Antartica, Cladosporium austrolitorale from coastal sea sand. Australia, Austroboletus yourkae on soil, Crepidotus innuopurpureus on dead wood, Curvularia stenotaphri from roots and leaves of Stenotaphrum secundatum and Thecaphora stajsicii from capsules of Oxalis radicosa. Belgium, Paraxerochrysium coryli (incl. Paraxerochrysium gen. nov.) from Corylus avellana. Brazil, Calvatia nordestina on soil, Didymella tabebuiicola from leaf spots on Tabebuia aurea, Fusarium subflagellisporum from hypertrophied floral and vegetative branches of Mangifera indica and Microdochium maculosum from living leaves of Digitaria insularis. Canada, Cuphophyllus bondii from a grassland. Croatia, Mollisia inferiseptata from a rotten Laurus nobilis trunk. Cyprus, Amanita exilis on calcareous soil. Czech Republic, Cytospora hippophaicola from wood of symptomatic Vaccinium corymbosum. Denmark, Lasiosphaeria deviata on pieces of wood and herbaceous debris. Dominican Republic, Calocybella goethei among grass on a lawn. France (Corsica), Inocybe corsica on wet ground. France (French Guiana), Trechispora patawaensis on decayed branch of unknown angiosperm tree and Trechispora subregularis on decayed log of unknown angiosperm tree. Germany, Paramicrothecium sambuci (incl. Paramicrothecium gen. nov.) on dead stems of Sambucus nigra. India, Aureobasidium microtermitis from the gut of a Microtermes sp. termite, Laccaria diospyricola on soil and Phylloporia tamilnadensis on branches of Catunaregam spinosa. Iran, Pythium serotinoosporum from soil under Prunus dulcis. Italy, Pluteus brunneovenosus on twigs of broadleaved trees on the ground. Japan, Heterophoma rehmanniae on leaves of Rehmannia glutinosa f. hueichingensis. Kazakhstan, Murispora kazachstanica from healthy roots of Triticum aestivum. Namibia, Caespitomonium euphorbiae (incl. Caespitomonium gen. nov.) from stems of an Euphorbia sp. Netherlands, Alfaria junci, Myrmecridium junci, Myrmecridium juncicola, Myrmecridium juncigenum, Ophioceras junci, Paradinemasporium junci (incl. Paradinemasporium gen. nov.), Phialoseptomonium junci, Sporidesmiella juncicola, Xenopyricularia junci and Zaanenomyces quadripartis (incl. Zaanenomyces gen. nov.), from dead culms of Juncus effusus, Cylindromonium everniae and Rhodoveronaea everniae from Evernia prunastri, Cyphellophora sambuci and Myrmecridium sambuci from Sambucus nigra, Kiflimonium junci, Sarocladium junci, Zaanenomyces moderatricis-academiae and Zaanenomyces versatilis from dead culms of Juncus inflexus, Microcera physciae from Physcia tenella, Myrmecridium dactylidis from dead culms of Dactylis glomerata, Neochalara spiraeae and Sporidesmium spiraeae from leaves of Spiraea japonica, Neofabraea salicina from Salix sp., Paradissoconium narthecii (incl. Paradissoconium gen. nov.) from dead leaves of Narthecium ossifragum, Polyscytalum vaccinii from Vaccinium myrtillus, Pseudosoloacrosporiella cryptomeriae (incl. Pseudosoloacrosporiella gen. nov.) from leaves of Cryptomeria japonica, Ramularia pararhabdospora from Plantago lanceolata, Sporidesmiella pini from needles of Pinus sylvestris and Xenoacrodontium juglandis (incl. Xenoacrodontium gen. nov. and Xenoacrodontiaceae fam. nov.) from Juglans regia. New Zealand, Cryptometrion metrosideri from twigs of Metrosideros sp., Coccomyces pycnophyllocladi from dead leaves of Phyllocladus alpinus, Hypoderma aliforme from fallen leaves Fuscopora solandri and Hypoderma subiculatum from dead leaves Phormium tenax. Norway, Neodevriesia kalakoutskii from permafrost and Variabilispora viridis from driftwood of Picea abies. Portugal, Entomortierella hereditatis from a biofilm covering a deteriorated limestone wall. Russia, Colpoma junipericola from needles of Juniperus sabina, Entoloma cinnamomeum on soil in grasslands, Entoloma verae on soil in grasslands, Hyphodermella pallidostraminea on a dry dead branch of Actinidia sp., Lepiota sayanensis on litter in a mixed forest, Papiliotrema horticola from Malus communis, Paramacroventuria ribis (incl. Paramacroventuria gen. nov.) from leaves of Ribes aureum and Paramyrothecium lathyri from leaves of Lathyrus tuberosus. South Africa, Harzia combreti from leaf litter of Combretum collinum ssp. sulvense, Penicillium xyleborini from Xyleborinus saxesenii, Phaeoisaria dalbergiae from bark of Dalbergia armata, Protocreopsis euphorbiae from leaf litter of Euphorbia ingens and Roigiella syzygii from twigs of Syzygium chordatum. Spain, Genea zamorana on sandy soil, Gymnopus nigrescens on Scleropodium touretii, Hesperomyces parexochomi on Parexochomus quadriplagiatus, Paraphoma variabilis from dung, Phaeococcomyces kinklidomatophilus from a blackened metal railing of an industrial warehouse and Tuber suaveolens in soil under Quercus faginea. Svalbard and Jan Mayen, Inocybe nivea associated with Salix polaris. Thailand, Biscogniauxia whalleyi on corticated wood. UK, Parasitella quercicola from Quercus robur. USA, Aspergillus arizonicus from indoor air in a hospital, Caeliomyces tampanus (incl. Caeliomyces gen. nov.) from office dust, Cippumomyces mortalis (incl. Cippumomyces gen. nov.) from a tombstone, Cylindrium desperesense from air in a store, Tetracoccosporium pseudoaerium from air sample in house, Toxicocladosporium glendoranum from air in a brick room, Toxicocladosporium losalamitosense from air in a classroom, Valsonectria portsmouthensis from air in men’s locker room and Varicosporellopsis americana from sludge in a water reservoir. Vietnam, Entoloma kovalenkoi on rotten wood, Fusarium chuoi inside seed of Musa itinerans, Micropsalliota albofelina on soil in tropical evergreen mixed forests and Phytophthora docyniae from soil and roots of Docynia indica. Morphological and culture characteristics are supported by DNA barcodes.
Citation: Crous PW, Osieck ER, Jurjević Ž, et al. 2021. Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. https://doi.org/10.3767/persoonia.2021.47.06.
Keywords: ITS nrDNA barcodes, LSU, new taxa, systematics
Acknowledgments
P.R. Johnston thanks J. Sullivan (Lincoln University) for the habitat image of Kowai Bush, Duckchul Park (Manaaki Whenua – Landcare Research) for the DNA sequencing, and the New Zealand Department of Conservation for permission to collect the specimens; this research was supported through the Manaaki Whenua – Landcare Research Biota Portfolio with funding from the Science and Innovation Group of the New Zealand Ministry of Business, Innovation and Employment. V. Hubka was supported by the Czech Ministry of Health (grant number NU21-05-00681), and is grateful for the support from the Japan Society for the Promotion of Science – grant-in-aid for JSPS research fellow (grant no. 20F20772). K. Glässnerová was supported by the Charles University Grant Agency (grant No. GAUK 140520). J. Trovão and colleagues were financed by FEDER- Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 – Operational Programme for Competitiveness and Internationalisation (POCI), and by Portuguese funds through FCT – Fundação para a Ciência e a Tecnologia in the framework of the project POCI-01-0145-FEDER-PTDC/EPH-PAT/3345/2014. This work was carried out at the R&D Unit Centre for Functional Ecology – Science for People and the Planet (CFE), with reference UIDB/04004/2020, financed by FCT/MCTES through national funds (PIDDAC). J. Trovão was also supported by POCH – Programa Operacional Capital Humano (co-funding by the European Social Fund and national funding by MCTES), through a ‘FCT – Fundação para a Ciência e Tecnologia’ PhD research grant (SFRH/BD/132523/2017). D. Haelewaters acknowledges support from the Research Foundation – Flanders (Junior Postdoctoral Fellowship 1206620N). M. Loizides and colleagues are grateful to Y. Cherniavsky for contributing collections AB A12-058-1 and AB A12-058-2, and Á. Kovács and B. Kiss for their help with molecular studies of these specimens. C. Zmuda is thanked for assisting with the collection of ladybird specimens infected with Hesperomyces parexochomi. A.V. Kachalkin and colleagues were supported by the Russian Science Foundation (grant No. 19-74-10002). The study of A.M. Glushakova was carried out as part of the Scientific Project of the State Order of the Government of Russian Federation to Lomonosov Moscow State University No. 121040800174-6. S. Nanu acknowledges the Kerala State Council for Science, Technology and Environment (KSCSTE) for granting a research fellowship and is grateful to the Chief Conservator of Forests and Wildlife for giving permission to collect fungal samples. A. Bañares and colleagues thank L. Monje and A. Pueblas of the Department of Drawing and Scientific Photography at the University of Alcalá for their help in the digital preparation of the photographs, and J. Rejos, curator of the AH herbarium for his assistance with the specimens examined in the present study. The research of V. Antonín received institutional support for long-term conceptual development of research institutions provided by the Ministry of Culture (Moravian Museum, ref. MK000094862). The studies of E.F. Malysheva, V.F. Malysheva, O.V. Morozova, and S.V. Volobuev were carried out within the framework of a research project of the Komarov Botanical Institute RAS, St Petersburg, Russia (AAAA-A18-118022090078-2) using equipment of its Core Facility Centre ‘Cell and Molecular Technologies in Plant Science’.The study of A.V. Alexandrova was carried out as part of the Scientific Project of the State Order of the Government of Russian Federation to Lomonosov Moscow State University No. 121032300081-7. The Kits van Waveren Foundation (Rijksherbariumfonds Dr E. Kits van Waveren, Leiden, Netherlands) contributed substantially to the costs of sequencing and travelling expenses for M.E. Noordeloos. The work of B. Dima was partly supported by the ÚNKP-20-4 New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund. The work of L. Nagy was supported by the ‘Momentum’ program of the Hungarian Academy of Sciences (contract No. LP2019-13/2019 to L.G.N.). G.A. Kochkina and colleagues acknowledge N. Demidov for the background photograph, and N. Suzina for the SEM photomicrograph. The research of C.M. Visagie and W.J. Nel was supported by the National Research Foundation grant no 118924 and SFH170610239162. C. Gil-Durán acknowledges Agencia Nacional de Investigación y Desarrollo, Ministerio de Ciencia, Tecnología, Conocimiento e Innovación, Gobierno de Chile, for grant ANID – Fondecyt de Postdoctorado 2021 – N° 3210135. R. Chávez and G. Levicán thank DICYT-USACH and acknowledges the grants INACH RG_03-14 and INACH RT_31-16 from the Chilean Antarctic Institute, respectively. S. Tiwari and A. Baghela would like to acknowledge R. Avchar and K. Balasubramanian from the Agharkar Research Institute, Pune, Maharashtra for helping with the termite collection. S. Tiwari is also thankful to the University Grants Commission, Delhi (India) for a junior research fellowship (827/(CSIR-UGC NET DEC.2017)). R. Lebeuf and I. Saar thank D. and H. Spencer for collecting and photographing the holotype of C. bondii, and R. Smith for photographing the habitat. A. Voitk is thanked for helping with the colour plate and review of the manuscript, and the Foray Newfoundland and Labrador for providing the paratype material. I. Saar was supported by the Estonian Research Council (grant PRG1170) and the European Regional Development Fund (Centre of Excellence EcolChange). M.P.S. Câmara acknowledges the ‘Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq’ for the research productivity fellowship, and financial support (Universal number 408724/2018-8). W.A.S. Vieira acknowledges the ‘Coordenação de Aperfeiçoamento Pessoal de Ensino Superior – CAPES’ and the ‘Programa Nacional de Pós-Doutorado/CAPES – PNPD/CAPES’ for the postdoctoral fellowship. A.G.G. Amaral acknowledges CNPq, and A.F. Lima and I.G. Duarte acknowledge CAPES for the doctorate fellowships. F. Esteve-Raventós and colleagues were financially supported by FEDER/Ministerio de Ciencia, Innovación y Universidades – Agencia Estatal de Investigación (Spain)/ Project CGL2017-86540-P. The authors would like to thank L. Hugot and N. Suberbielle (Conservatoire Botanique National de Corse, Office de l’Environnement de la Corse, Corti) for their help. The research of E. Larsson is supported by The Swedish Taxonomy Initiative, SLU Artdatabanken, Uppsala. Financial support was provided to R.J. Ferreira by the National Council for Scientific and Technological Development (CNPq), and to I.G. Baseia, P.S.M. Lúcio and M.P. Martín by the National Council for Scientific and Technological Development (CNPq) under CNPq-Universal 2016 (409960/2016-0) and CNPq-visiting researcher (407474/2013-7). J. Cabero and colleagues wish to acknowledge A. Rodríguez for his help to describe Genea zamorana, as well as H. Hernández for sharing information about the vegetation of the type locality. S. McMullan-Fisher and colleagues acknowledge K. Syme (assistance with illustrations), J. Kellermann (translations), M. Barrett (collection, images and sequences), T. Lohmeyer (collection and images) and N. Karunajeewa (for prompt accessioning). This research was supported through funding from Australian Biological Resources Study grant (TTC217-06) to the Royal Botanic Gardens Victoria. The research of M. Spetik and co-authors was supported by project No. CZ.02.1.01/0.0/0.0/16_017/0002334. N. Wangsawat and colleagues were partially supported by NRCT and the Royal Golden Jubilee Ph.D. programme, grant number PHD/0218/2559. They are thankful to M. Kamsook for the photograph of the Phu Khiao Wildlife Sanctuary and P. Thamvithayakorn for phylogenetic illustrations. The study by N.T. Tran and colleagues was funded by Hort Innovation (Grant TU19000). They also thank the turf growers who supported their surveys and specimen collection. N. Matočec, I. Kušan, A. Pošta, Z. Tkalčec and A. Mešić thank the Croatian Science Foundation for their financial support under the project grant HRZZ-IP-2018-01-1736 (ForFungiDNA). A. Pošta thanks the Croatian Science Foundation for their support under the grant HRZZ-2018-09-7081. A. Morte is grateful to Fundación Séneca – Agencia de Ciencia y Tecnología de la Región de Murcia (20866/PI/18) for financial support. The research of G. Akhmetova, G.M. Kovács, B. Dima and D.G. Knapp was supported by the National Research, Development and Innovation Office, Hungary (NKFIH KH-130401 and K-139026), the ELTE Thematic Excellence Program 2020 supported by the National Research, Development and Innovation Office (TKP2020-IKA-05) and the Stipendium Hungaricum Programme. The support of the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and the Bolyai+ New National Excellence Program of the Ministry for Innovation and Technology to D.G. Knapp is highly appreciated. F.E. Guard and colleagues are grateful to the traditional owners, the Jirrbal and Warungu people, as well as L. and P. Hales, Reserve Managers, of the Yourka Bush Heritage Reserve. Their generosity, guidance, and the opportunity to explore the Bush Heritage Reserve on the Einasleigh Uplands in far north Queensland is greatly appreciated. The National Science Foundation (USA) provided funds (DBI#1828479) to the New York Botanical Garden for a scanning electron microscope used for imaging the spores. V. Papp was supported by the ÚNKP-21-5 New National Excellence Program of the Ministry for Innovation and Technology from the National Research, Development and Innovation Fund of Hungary. A.N. Miller thanks the WM Keck Center at the University of Illinois Urbana – Champaign for sequencing Lasiosphaeria deviata. J. Pawłowska acknowledges support form National Science Centre, Poland (grant Opus 13 no 2017/25/B/NZ8/00473). The research of T.S. Bulgakov was carried out as part of the State Research Task of the Subtropical Scientific Centre of the Russian Academy of Sciences (Theme No. 0492-2021-0007). K. Bensch (Westerdijk Fungal Biodiversity Institute, Utrecht) is thanked for correcting the spelling of various Latin epithets.
REFERENCES
- Abad ZG, Burgess TI, Bienapfl JC, et al. 2019. IDphy: Molecular and morphological identification of Phytophthora based on the types. USDA APHIS PPQ S&T Beltsville Lab, USDA APHIS PPQ S&T ITP, Centre for Phytophthora Science and Management, and World Phytophthora Collection. [Google Scholar]
- Abdollahzadeh J, Groenewald JZ, Coetzee MPA, et al. 2020. Evolution of lifestyles in Capnodiales. Studies in Mycology 95: 381–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado P, Cabero J, Moreno G, et al. 2014. Species diversity of Genea (Ascomycota, Pezizales) in Europe. Ascomycete.org 6: 41–51. [Google Scholar]
- Alvarado P, Cabero J, Moreno G, et al. 2016. Phylogenetic overview of the genus Genea (Pezizales, Ascomycota) with an emphasis on European taxa. Mycologia 108: 441–456. [DOI] [PubMed] [Google Scholar]
- Alvarado P, Pérez J-B, Van Vooren N, et al. 2020. Genea coronata (Pyronemataceae, Pezizales), a cryptic new species in a highly polymorphic genus. Sydowia 72: 231–242. [Google Scholar]
- Anonymous. 1969. Flora of British Fungi – Colour Identification Chart. Her Majesty’s Stationery Office, Edinburgh. [Google Scholar]
- Antonín V, Noordeloos M. 2010. A monograph of marasmioid and collybioid fungi in Europe. IHW Verlag. [Google Scholar]
- Arzanlou A, Groenewald JZ, Gams W, et al. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp MM, Verkley GJM, De Gruyter J, et al. 2009. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 101: 363–382. [DOI] [PubMed] [Google Scholar]
- Bandini D, Oertel B, Ploch S, et al. 2019. Revision of some central European species of Inocybe (Fr.: Fr.) Fr. subgenus Inocybe, with description of five new species. Mycological Progress 18: 247–294. [Google Scholar]
- Bandini D, Sesli E, Oertel B, et al. 2020. Inocybe antoniniana, a new species of Inocybe section Marginatae with nodulose spores. Sydowia 72: 95–106. [Google Scholar]
- Bao DF, Wanasinghe DN, Luo ZL, et al. 2019. Murispora aquatica sp. nov. and Murispora fagicola, a new record from freshwater habitat in China. Phytotaxa 416: 1–13. [Google Scholar]
- Baral H-O. 1987. Lugol’s solution / IKI versus Melzer’s reagent: hemiamyloidity, a universal feature of the ascus wall. Mycotaxon 29: 399–450. [Google Scholar]
- Bates ST, Roberson RW, Desjardin DE. 2009. Arizona gasteroid fungi I: Lycoperdaceae (Agaricales, Basidiomycota). Fungal Diversity 37: 153–207. [Google Scholar]
- Bensch K, Groenewald JZ, Braun U, et al. 2015. Common but different: The expanding realm of Cladosporium. Studies in Mycology 82: 23–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch K, Groenewald JZ, Meijer M, et al. 2018. Cladosporium species in indoor environments. Studies in Mycology 89: 177–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkeley MJ. 1845. Decades of Fungi: Australian Fungi. The London Journal of Botany 4: 42–73. [Google Scholar]
- Bertault R. 1965. Amanites du Maroc. Bulletin trimestriel de la Société mycologique de France 81: 345–371. [Google Scholar]
- Bezerra JDP, Sandoval-Denis M, Paiva LM, et al. 2017. New endophytic Toxicocladosporium species from cacti in Brazil, and description of Neocladosporium gen. nov. IMA Fungus 8: 77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien S, Kraus C, Damm U. 2020. Novel collophorina-like genera and species from Prunus trees and vineyards in Germany. Persoonia 45: 46–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerema GH, De Gruyer J, Noordeloos ME, et al. 2004. Phoma identification manual. Differentiation of specific and intra-specific and intra-specific taxa in culture. CABI Publishing, Wallingford, UK. [Google Scholar]
- Bose T, Wingfield MJ, Roux J, et al. 2021. Phytophthora species associated with roots of native and non-native trees in natural and managed forests Microbial Ecology 81: 122–133. [DOI] [PubMed] [Google Scholar]
- Braun U. 1998. A monograph of Cercosporella, Ramularia and allied genera (phytopathogenic hyphomycetes), Vol. 2, IHW-Verlag, Eching, Munich, Germany. [Google Scholar]
- Buisman CJ. 1927. Root rots caused by Phycomycetes. Mededelingen uit het Phytopathologisch laboratorium Willie Commelin Scholten 11: 1–65. [Google Scholar]
- Bukharova NV, Zmitrovich IV. 2014. Aphyllophoroid fungi of the Bastak Reserve. Mikologiya i fitopatologiya 48: 343–354. [Google Scholar]
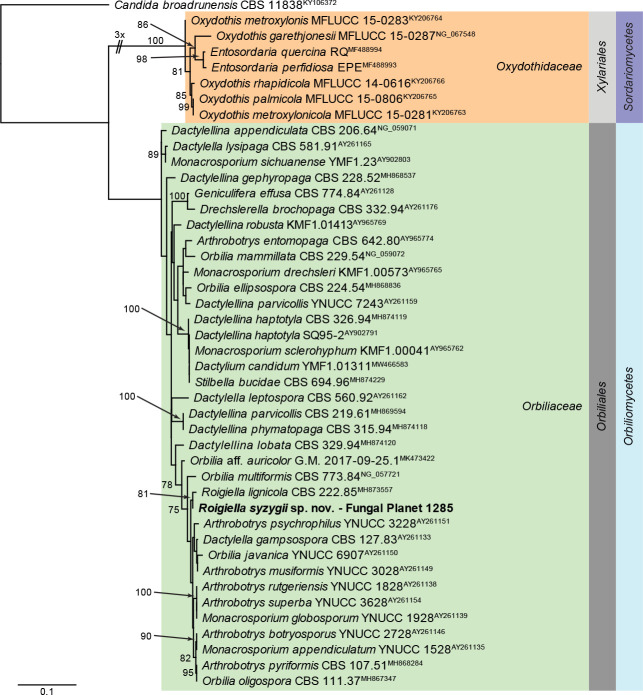
- Castañeda-Ruiz RF. 1984. Nuevos taxones de Deuteromycotina: Arnoldiella robusta gen. et sp. nov.; Roigiella lignicola gen. et sp. nov.; Sporidesmium pseudolmediae sp. nov. y Thozetella havanensis sp. nov. Revista del Jardín Botánico Nacional Universidad de la Habana 5: 57–87. [Google Scholar]
- Castañeda-Ruiz RF, Gams W, Saikawa M. 1997. Three new conidial fungi (hyphomycetes) from Cuba. Nova Hedwigia 64: 473–483. [Google Scholar]
- Chen C, Verkley GJM, Sun G, et al. 2016. Redefining common endophytes and plant pathogens in Neofabraea, Pezicula, and related genera. Fungal Biology 120: 1291–1322. [DOI] [PubMed] [Google Scholar]
- Chen Q, Hou LW, Duan WJ, et al. 2017. Didymellaceae revisited. Studies in Mycology 87: 105–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jiang JR, Zhang GZ, et al. 2015. Resolving the Phoma enigma. Studies in Mycology 82: 137–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contu M. 1985a ‘1984’. Novitates (2). Documents Mycologiques 14: 26. [Google Scholar]
- Contu M. 1985b. Amanita mairei Foley sensu Foley. A. mairei sensu Bertault: Nome nuovo per la specie di Bertault: Amanita bertaultii sp. nov. Funghie Ambiente 1: 11–13. [Google Scholar]
- Contu M. 2000. Chiave per la determinazione delle specie europee del genere Amanita sez. Vaginatae. Bollettino del gruppo micologico G. Bresadola 43: 233–240. [Google Scholar]
- Contu M, Pacioni G. 1998. Amanita cistetorum and Psathyrella liciosae, two new Mediterranean species. Mycotaxon 69: 437–446. [Google Scholar]
- Corriol G. 2003. Pluteus diverticulatus sp. nov., une espece nouvelle de la sous-section Mixtini. Bulletin Trimestriel de la Societe Mycologique de France 119: 231–244. [Google Scholar]
- Corriol G, Moreau PA, Bellanger JM. 2017. Calocybe hymenoderma sp. nov., Calocybella juncicola comb. nov. et les contours taxinomiques de la section Rugosomyces. Errotari 14: 35–46. [Google Scholar]
- Crandall BS. 1947. A new Phytophthora causing root and collar rot of cinchona in Peru. Mycologia 39: 218–223. [PubMed] [Google Scholar]
- Crous PW, Braun U, McDonald BA, et al. 2020a. Redefining genera of cereal pathogens: Oculimacula, Rhynchosporium and Spermospora. Fungal Systematics and Evolution 7: 67–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Wingfield MJ, et al. 2009. Phylogeny and taxonomy of obscure genera of microfungi. Persoonia 22: 139–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, et al. 2019a. Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. 2021a. Fungal Planet description sheets: 1182–1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Mansilla JP, et al. 2004. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. Studies in Mycology 50: 195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. 2021b. Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-ard JJ, Wingfield MJ, et al. 2018a. Fungal Planet description sheets: 785–867. Persoonia 41: 238–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, et al. 2019b. New and Interesting Fungi. 2. Fungal Systematics and Evolution 3: 57–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. 2018b. New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014a. Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, et al. 2011. Fungal Planet description sheets: 92–106. Persoonia 27: 130–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Christensen M, et al. 2012. How important are conidial appendages? Persoonia 28: 126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2017. Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Cheewangkoon R, et al. 2019c. Foliar pathogens of eucalypts. Studies in Mycology 94: 125–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi Y-H, et al. 2020b. Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, le Roux JJ, et al. 2015. Fungal Planet description sheets: 371–399. Persoonia 35: 264–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, et al. 2019d. Fungal Planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, et al. 2016. Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, et al. 2014b. Fungal Planet description sheets 281–319. Persoonia 33: 212–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, et al. 2020c. New and interesting fungi 3. Fungal Systematics and Evolution 6: 157–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gruyter J, Noordeloos ME. 1992. Contributions towards a monograph of Phoma (Coelomycetes) – I. 1. Section Phoma: Taxa with very small conidia in vitro. Persoonia 15: 71–92. [Google Scholar]
- De Gruyter J, Noordeloos ME, Boerema GH. 1993. Contributions towards a monograph of Phoma (Coelomycetes) – I. 2. Section Phoma: Additional taxa with very small conidia and taxa with conidia up to 7 μm long. Persoonia 15: 369–400. [Google Scholar]
- De Hoog GS. 1977. Rhinocladiella and allied genera. Studies in Mycology 15: 1–140. [Google Scholar]
- De Hoog GS, Mayser P, Haase G, et al. 2000. A new species, Phialophora europaea, causing superficial infections in humans. Mycoses 43: 409–416. [DOI] [PubMed] [Google Scholar]
- Denk T, Grimm GW. 2010. The oaks of western Eurasia: Traditional classifications and evidence from two nuclear markers. Taxon 59: 351–366. [Google Scholar]
- Diederich P, Braun U. 2009. First lichenicolous record of Acremonium hypholomatis (anamorphic Ascomycota). Bulletin de la Société des naturalistes luxembourgeois 110: 97–99. [Google Scholar]
- Domsch KH, Gams W, Anderson TH. 1993. Compendium of soil fungi. Reprinted by IHW-Verlag with supplement. [Google Scholar]
- Domsch KH, Gams W, Anderson T-H. 2007. Compendium of soil fungi. 2nd ed. IHW-Verlag, Eching, Germany. [Google Scholar]
- Dong W, Hyde KD, Jeewon R, et al. 2021. Towards a natural classification of annulatascaceae-like taxa II: introducing five new genera and eighteen new species from freshwater. Mycosphere 12: 1–88. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, et al. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecoluar Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JX, Wu WP, Liu XZ. 2007. Dinemasporium (coelomycetes). Fungal Diversity 26: 205–218. [Google Scholar]
- Edler D, Klein J, Antonelli A, et al. 2020. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12: 373–377. [Google Scholar]
- Egidi E, De Hoog GS, Isola D, et al. 2014. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fungal Diversity 65: 127–165. [Google Scholar]
- Ekanayaka AH, Hyde KD, Gentekaki E, et al. 2019. Preliminary classification of Leotiomycetes. Mycosphere 10: 310–489. [Google Scholar]
- Esteve-Raventós F, Moreno G, Bizio E, et al. 2015. Inocybe flavobrunnescens, a new species in section Marginatae. Mycological Progress 14: 14. [Google Scholar]
- Farr DF, Rossman AY. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Retrieved October 14, 2021, from https://nt.ars-grin.gov/fungaldatabases/. [Google Scholar]
- Fassatiová O. 1986. Moulds and filamentous fungi in technical microbiology. Elsevier, Amsterdam. [Google Scholar]
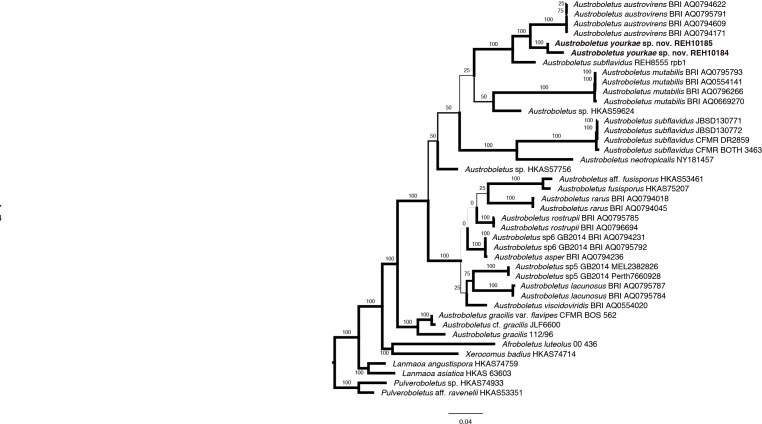
- Fechner N, Bonito G, Bougher NL, et al. 2017. New species of Austroboletus (Boletaceae) in Australia. Mycological Progress 16: 769–775. [Google Scholar]
- Feng P, Lu Q, Najafzadeh MJ, et al. 2014. Cyphellophora and its relatives in Phialophora biodiversity and possible role in human infection. Fungal Diversity 65: 17–45. [Google Scholar]
- Fries E. 1821. Systema mycologicum 1. Ex officina Berlingiana, Lund, Sweden. [Google Scholar]
- Gams W. 1971. Cephalosporium-artige Schimmelpilze (Hyphomycetes), Fischer, Stuttgart, Germany. [Google Scholar]
- Gams W. 1975. Cephalosporium-like Hyphomycetes: Some tropical species. Transactions of the British Mycological Society 64: 389–404. [Google Scholar]
- Gams W. 1976. Some new or noteworthy species of Mortierella. Persoonia 9: 111–140. [Google Scholar]
- Giraldo A, Gené J, Sutton DA, et al. 2015. Phylogeny of Sarocladium (Hypocreales). Persoonia 34: 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves MFM, Santos L, Silva BMV, et al. 2019. Biodiversity of Penicillium species from marine environments in Portugal and description of Penicillium lusitanum sp. nov., a novel species isolated from sea water. International Journal of Systematic and Evolutionary Microbiology 69: 3014–3021. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Gräfenhan T, Schroers H-J, Nirenberg HI, et al. 2011. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Studies in Mycology 68: 79–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryzenhout M, Tarigan M, Clegg PA, et al. 2010. Cryptometrion aestuescens gen. sp. nov. (Cryphonectriaceae) pathogenic to Eucalyptus in Indonesia. Australasian Plant Pathology 39: 161–169. [Google Scholar]
- Guindon S, Lethiec F, Duroux P. et al. 2010. PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research 33: W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán-Dávalos L, Pradeep CK, Vrinda KB, et al. 2017. A new stipitate species of Crepidotus from India and Thailand, with notes on other tropical species. Mycologia 109: 804–814. [DOI] [PubMed] [Google Scholar]
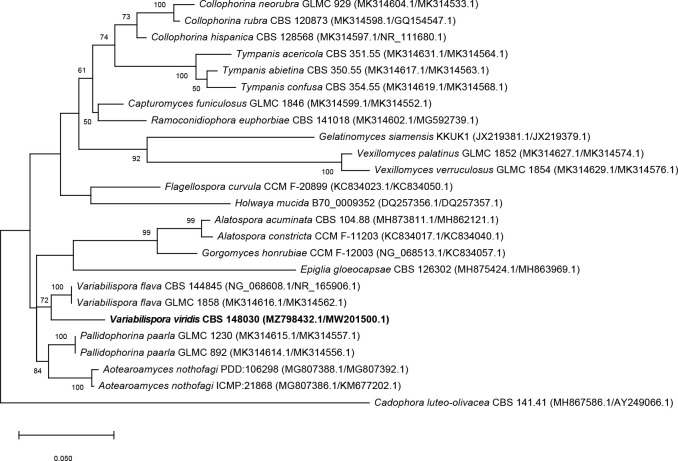
- Haelewaters D, De Kesel A. 2020. Checklist of thallus-forming Laboulbeniomycetes from Belgium and the Netherlands, including Hesperomyces halyziae and Laboulbenia quarantenae spp. nov. MycoKeys 71: 23–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haelewaters D, De Kesel A, Pfister DH. 2018. Integrative taxonomy reveals hidden species within a common fungal parasite of ladybirds. Scientific Reports 8: 15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Halling RE. 1996. Notes on Collybia V. Gymnopus section Levipedes in tropical South America, with comments on Collybia. Brittonia 48: 487–494. [Google Scholar]
- Hanss J-M, Moreau P-A. 2020. Une Reìvision des Amanites «vagineìes» (Amanita sect. Vaginatae) en Europe, 1re partie: quelques Amanites argenteìes. Bulletin de la Socieìteì mycologique de France 133: 67–141. [Google Scholar]
- Hashimoto A, Sato G, Matsuda T, et al. 2015. Taxonomic revision of Pseudolachnea and Pseudolachnella and establishment of Neopseudolachnella and Pseudodinemasporium gen. nov. Mycologia 107: 383–408. [DOI] [PubMed] [Google Scholar]
- Hawksworth DL. 1972. A large-spored species of Acremonium sect. Nectrioidea. Transactions of the British Mycological Society 58: 510–512. [Google Scholar]
- Hawksworth DL. 1979. The lichenicolous hyphomycetes. Bulletin of the British Museum for Natural History 6: 183–300. [Google Scholar]
- He M-Q, Hyde KD, Ratchadawan C, et al. 2020. Two new species of Micropsalliota (Agaricaceae/Agaricales) from Thailand. Phytotaxa 453: 140–141. [Google Scholar]
- Heinemann P, Leelavathy KM. 1991. The genus Micropsalliota (Agaricaceae) in Kerala State, India. Mycological Research 95: 343–344. [Google Scholar]
- Heinemann P, Little Flower S. 1983. Micropsalliota de Kerala (Inde). Bulletin du Jardin Botanique National de Belgique 53: 75–84. [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Crous PW, et al. 2016a. Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia 36: 57–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Elliott ML, et al. 2016b. Take-all or nothing. Studies in Mycology 83: 19–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp AL, Manos PS, Hahn M, et al. 2020. Genomic landscape of the global oak phylogeny. New Phytologist 226: 1198–1212. [DOI] [PubMed] [Google Scholar]
- Hjortstam K, Ryvarden L. 2007. Checklist of corticioid fungi (Basidiomycotina) from the tropics, subtropics and the southern hemisphere. Synopsis Fungorum 22: 27–146. [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, et al. 2018. UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubová-Jechová V. 1974. A revision of the genus Olpitrichum Atk. Folia Geobotanica et Phytotaxonomica 9: 425–432. [Google Scholar]
- Hongo T. 1971. Notulae mycologicae. Mem Shiga University 21: 62–68. [Google Scholar]
- Horak E. 1976. On cuboid spored species of Entoloma. Sydowia 28: 171–236. [Google Scholar]
- Horak E. 1977. Additions to ‘On cuboid spored species of Entoloma’. Sydowia 29: 289–299. [Google Scholar]
- Horak E. 1978 ‘1977’. Crepidotus episphaeria and related species from the Southern Hemisphere. Berichte der Schweizerischen Botanischen Gesellschaft 87: 227–235. [Google Scholar]
- Horak E. 2018. Agaricales (Basidiomycota) of New Zealand 2. Brown spored genera p.p. Crepidotus, Flammulaster, Inocybe, Phaeocollybia, Phaeomarasmius, Pleuroflammula, Pyrrhoglossum, Simocybe, Tubaria and Tympanella. Fungi of New Zealand Nga Hekaheka o Aotearoa Volume 6. Westerdijk Biodiversity Series 16: 1–205. Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. [Google Scholar]
- Hou C-L, Lin Y-R, Piepenbring M. 2005. Species of Rhytismataceae on needles of Juniperus spp. from China. Canadian Journal of Botany 83: 37–46. [Google Scholar]
- Hou LW, Groenewald JZ, Pfenning LH, et al. 2020a. The phoma-like dilemma. Studies in Mycology 96: 309–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou LW, Hernandez-Restrepo M, Groenewald JZ, et al. 2020b. Citizen science project reveals high diversity in Didymellaceae (Pleosporales, Dothideomycetes). MycoKeys 65: 49–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Kocsube S, Visagie CM, et al. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
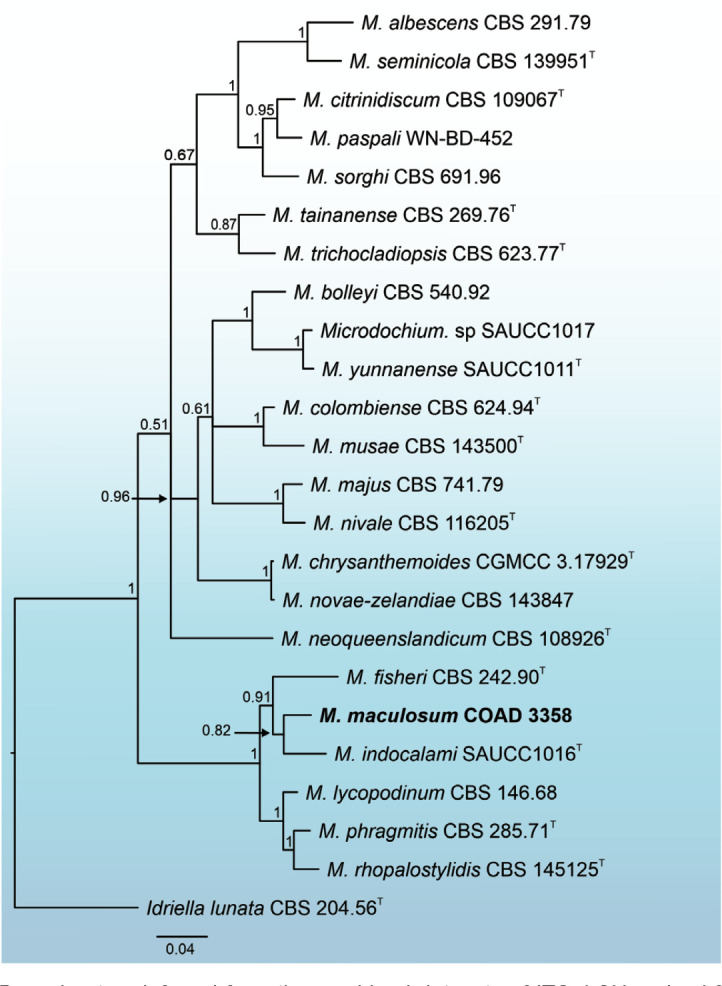
- Huang S, Xia J, Zhang X, et al. 2020. Two new species of Microdochium from Indocalamus longiauritus in south-western China. MycoKeys 72: 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Dudová Z, Kubátová A, et al. 2017. Taxonomic novelties in Aspergillus section Fumigati: A. tasmanicus sp. nov., induction of sexual state in A. turcosus and overview of related species. Plant Systematics and Evolution 303: 787–806. [Google Scholar]
- Hubka V, Peterson SW, Frisvad JC, et al. 2013. Aspergillus waksmanii sp. nov. and Aspergillus marvanovae sp. nov., two closely related species in section Fumigati. International Journal of Systematic and Evolutionary Microbiology 63: 783–789. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Huson DH, Kloepper TH. 2005. Computing recombination networks from binary sequences. Bioinformatics, 21: ii159–ii165. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, et al. 2020. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. [Google Scholar]
- Hyde KD, Norphanphoun C, Abreu VP, et al. 2017. Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87: 1–235. [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R. et al. 2019. Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. [Google Scholar]
- Hymphries Z, Seifert KA, Hirooka Y, et al. 2017. A new family and genus in Dothideales for Aureobasidium-like species isolated from house dust. IMA Fungus 8: 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JAG. 1991. Phytophthora macrochlamydospora, a new species from Australia. Mycologia 83: 517–519. [Google Scholar]
- Jiang H-B, Hyde KD, Yang E-F, et al. 2021. Morphological and phylogenetic appraisal of Ophioceras (Ophioceraceae, Magnaporthales). PLoS ONE 16: e0253853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Fan X, Tian C, et al. 2020. Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia 112: 267–292. [DOI] [PubMed] [Google Scholar]
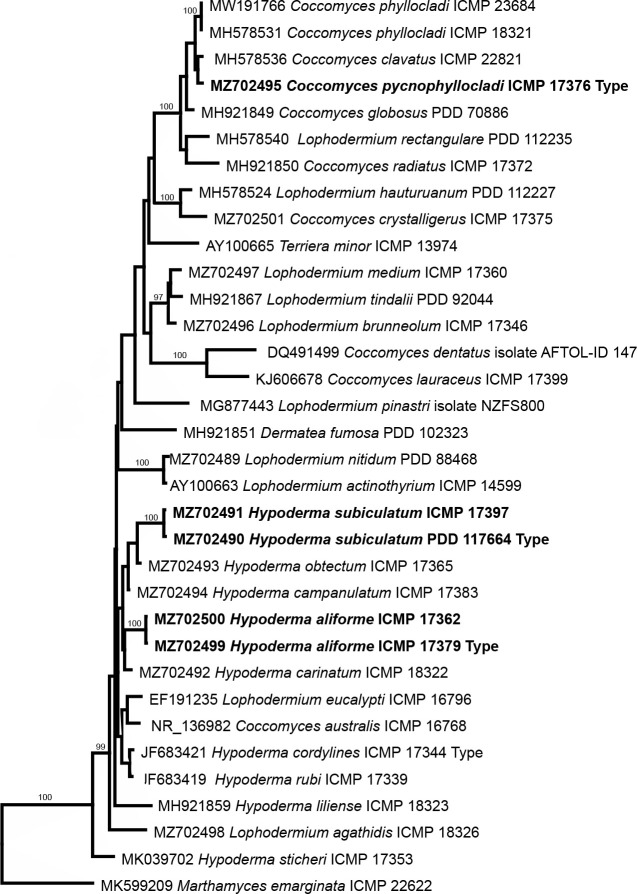
- Johnston PR. 1986. Rhytismataceae in New Zealand 1. Some foliicolous species of Coccomyces de Notaris and Propolis (Fries) Corda. New Zealand Journal of Botany 24: 89–124. [Google Scholar]
- Johnston PR. 1994. Ascospore sheaths of some Coccomyces, Hypoderma, and Lophodermium species (Rhytismataceae). Mycotaxon 52: 221–239. [Google Scholar]
- Johnston PR, Gamundi IJ. 2000. Torrendiella (Ascomycota, Helotiales) on Nothofagus. New Zealand Journal of Botany 38: 493–513. [Google Scholar]
- Ju Y-M, Rogers JD. 2001. New and interesting Biscogniauxia taxa, with a key to the world species. Mycological Research 9: 1123–1133. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TFK, et al. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karstedt F, Baroni TJ, Largent DL, et al. 2019. Phylogenetic and morphological analyses of species of the Entolomataceae (Agaricales, Basidiomycota) with cuboid basidiospores. Phytotaxa 391: 001–027. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalabuda TV. 1973. Fungi of the genus Mortierella Coemans. Institut mikrobiologii (Akademiia nauk SSSR). [Google Scholar]
- Kirk PM. 1982. New or interesting microfungi VI. Sporidesmiella gen. nov. (hyphomycetes). Transactions of the British Mycological Society 79: 479–489. [Google Scholar]
- Klaubauf S, Tharreau D, Fournier E, et al. 2014. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Studies in Mycology 79: 85–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DG, Kovács GM, Zajta E, et al. 2015. Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia 35: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Courtecuisse R. 2000. Two new species of Inocybe, section Marginatae (Agaricales, Cortinariaceae) from Japan. Mycoscience 41: 161–166. [Google Scholar]
- Kornerup A, Wanscher JH. 1967. Methuen Handbook of Colour. Methuen & Co Ltd, London, England. [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen Handbook of Colour, 3rd edn. Eyre Methuen, Ltd., London. [Google Scholar]
- Kornerup A, Wanscher JH. 1983. Methuen Handbook of Colour, 3rd edn. Eyre Methuen, Ltd., London. [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, et al. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinfomatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys A, Huhndorf SM, Miller AN. 2015. Coprophilous contributions to the phylogeny of Lasiosphaeriaceae and allied taxa within Sordariales (Ascomycota, Fungi). Fungal Diversity 70: 101–113. [Google Scholar]
- Kühner R. 1955. Compléments a la “Flore Analytique” VI) Inocybe goniosporés et Inocybe acystidiés. Espèces nouvelles ou critiques. Bulletin de la Société Mycologique de France 71: 279–311. [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kušan I, Matočec N, Antonić O, et al. 2014. Biogeographical variability and re-description of an imperfectly known species Hamatocanthoscypha rotundispora (Helotiales, Hyaloscyphaceae). Phytotaxa 170: 1–12. [Google Scholar]
- Kuyper TW. 1986. A revision of the genus Inocybe in Europe. I. Subgenus Inosperma and the smooth-spored species of subgenus Inocybe. Persoonia Supplement 3: 1–247. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Laraba I, McCormick SP, Vaughan MM, et al. 2021. Phylogenetic diversity, trichothecene potential, and pathogenicity within Fusarium sambucinum species complex. Plos One 16: e0245037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson KH. 1994. Poroid species in Trechispora and use of calcium oxalate crystals for species identification. Mycological Research 98: 1153–1172. [Google Scholar]
- Latha KD, Raj KA, Manimohan P. 2019. Laccaria violaceotincta: a new species from tropical India based on morphology and molecular phylogeny. Phytotaxa 392: 140–146. [Google Scholar]
- Latha KPD, Raj KNA, Thushara C, et al. 2016. Three new species of Calocybella from India based on morphology and molecular phylogeny. Phytotaxa 255: 133–143. [Google Scholar]
- Le Gal M, Mangenot MF. 1961. Contribution à l’étude des MollisioȈdées. IV. (3e série). Revue de Mycologie 26: 263–331. [Google Scholar]
- Lechat C, Fournier J. 2016. Varicosporellopsis, a new aquatic genus from South of France. Ascomycete.org 8: 96–100. [Google Scholar]
- Leonardi M, Furtado ANM, Comandini O, et al. 2020. Halimium as an ectomycorrhizal symbiont: new records and an appreciation of known fungal diversity Mycological Progress 19: 1495–1509. [Google Scholar]
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames. [Google Scholar]
- Lévesque CA, De Cock AWAM. 2004. Molecular phylogeny and taxonomy of the genus Pythium. Mycological Research 108: 1363–1383. [DOI] [PubMed] [Google Scholar]
- Li HY, Sun GY, Zhai XR, et al. 2012. Dissoconiaceae associated with sooty blotch and flyspeck on fruits in China and the United States. Persoonia 28: 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-X, He M-Q, Zhao R-L. 2021. Three new species of Micropsalliota (Agaricaceae, Agaricales) from China. Phytotaxa 491: 174. [Google Scholar]
- Liberta AE. 1973. The genus Trechispora (Basidiomycetes, Corticiaceae). Canadian Journal of Botany 51: 1871–1892. [Google Scholar]
- Lima CS, Pfenning LH, Costa SS, et al. 2009. A new Fusarium lineage within the Gibberella fujikuroi species complex is the main causal agent of mango malformation disease in Brazil. Plant Pathology 58: 33–42. [Google Scholar]
- Liu LL, Yang J, Liu NG, et al. 2019. Sporidesmium guizhouense sp. nov. (Sordariomycetes incertae sedis), a new species from a freshwater habitat in Guizhou Province, China. Phytotaxa 422: 144–156. [Google Scholar]
- Liu ZY, Plautz HL, Lumyong S, et al. 2016. Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87: 1–235. [Google Scholar]
- Loizides M, Bellanger J-M, Yiangou Y, et al. 2018. Preliminary phylogenetic investigations into the genus Amanita (Agaricales) in Cyprus, with a review of previous records and poisoning incidents. Documents Mycologiques XXXVII: 201–218. [Google Scholar]
- Lombard L, Houbraken J, Decock C, et al. 2016. Generic hyper-diversity in Stachybotriaceae. Persoonia 36: 156–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van der Merwe NA, Groenewald JZ, et al. 2015. Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe JL. 1966. Polyporaceae of North America. The genus Poria. New York State University College of Forestry Technical Publication 90. [Google Scholar]
- Lu YZ, Liu JK, Hyde KD, et al. 2018. A taxonomic reassessment of Tubeufiales based on multi-locus phylogeny and morphology. Fungal Diversity 92: 131–344. [Google Scholar]
- Ludwig E. 2007. Pilzkompendium (Eching), 2. Berlin. Ed. Fungicon. [Google Scholar]
- Lundquist N. 1972. Nordic Sordariaceae s. lat. Symbolae Botanicae Upsalienses 20: 1–374. [Google Scholar]
- Luo ZL, Hyde KD, Bhat DJ, et al. 2018. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress 17: 511–530. [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, et al. 2019. Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. [Google Scholar]
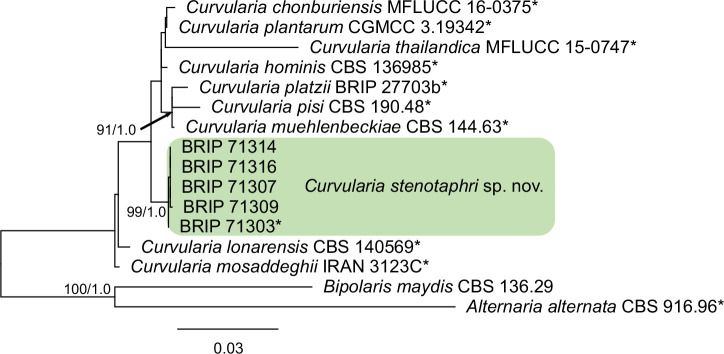
- Madrid H, da Cunha KC, Gené J, et al. 2014. Novel Curvularia species from clinical specimens. Persoonia 33: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoj KA, Vrinda KB, Pradeep CK. 2018. New and noteworthy crepidotoid agarics from India. Cryptogamie, Mycologie 39: 287–298. [Google Scholar]
- Maryani N, Sandoval-Denis M, Lombard L, et al. 2019. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 43: 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Felix Y, Guarro J, Cano-Lira JF, et al. 2018. Melanospora (Sordariomycetes, Ascomycota) and its relatives. MycoKeys 44: 81–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata JL, Hughes KW, Petersen RH. 2006. An investigation of /omphalotaceae (Fungi: Euagarics) with emphasis on the genus Gymnopus. Sydowia 58: 191–289. [Google Scholar]
- Matheny BP, Swenie RA. 2018. The Inocybe geophylla group in North America: a revision of the lilac species surrounding I. lilacina. Mycologia 110: 618–634. [DOI] [PubMed] [Google Scholar]
- McKenzie EHC, Pinnoi A, Wong MKM, et al. 2002. Two new hyaline Chalara species, and a key to species described since 1975. Fungal Diversity 11: 129–139. [Google Scholar]
- Meiras-Ottoni A, Larsson K-H, Gibertoni TB. 2021. Additions to Trechispora and the status of Scytinopogon (Trechisporales, Basidiomycota). Mycological Progress 20: 203–222. [Google Scholar]
- Menolli N, Jr, Asai T, Capelari M. 2010. Records and new species of Pluteus from Brazil based on morphological and molecular data. Mycology 1: 130–153. [Google Scholar]
- Miller AN, Huhndorf SM. 2004a. A natural classification of Lasiosphaeria based on nuclear LSU rDNA sequences. Mycological Research 108: 26–34. [DOI] [PubMed] [Google Scholar]
- Miller AN, Huhndorf SM. 2004b. Using phylogenetic species recognition to delimit species boundaries within Lasiosphaeria. Mycologia 96: 1106–1127. [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA: 1–8. [Google Scholar]
- Minh BQ, Schmidt H.A., Chernomor O, et al. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter DW. 1996. Colpoma juniperi. IMI descriptions of Fungi and bacteria No 1294. Mycopathologia 136: 159–161. [DOI] [PubMed] [Google Scholar]
- Misra PC, Srivastava P. 1976. A new species of Tetracoccosporium. Mycotaxon 4: 276–278. [Google Scholar]
- Morgan AP. 1890. North American Fungi. The Gasteromycetes. III. The Journal of the Cincinnati Society of Natural History 12: 163–172. [Google Scholar]
- Morozova OV, Alexandrova AV, Popov ES, et al. 2016. New data on the agaricoid basidiomycetes of the Tver Region. Novosti Sistematiki Nizshikh Rastenii. 50: 174–186. [In Russian.] [Google Scholar]
- Morozova OV, Popov ES, Kovalenko AE. 2012. Studies on mycobiota of Vietnam. I. Genus Entoloma: new records and new species. Mikologiya i fitopatologiya 46: 184–200. [Google Scholar]
- Munsell Color 1994. Soil Color Charts (revised edition). Macbeth Division of Kollmorgen Instruments Corporation. New Windsor. New York. USA. [Google Scholar]
- Muroi T, Udagawa S, Otani Y. 1987. Some coprophilous ascomycetes from Peru. In: Inoue H. (ed), Studies on Cryptogams in Southern Peru. Tokai University Press, Tokyo: 151–168. [Google Scholar]
- Nag-Raj TR, Kendrick WB. 1975. Monograph of Chalara and allied genera. Wilfrid Laurier University Press, Waterloo, Ontario, Canada. [Google Scholar]
- Nagy LG, Petkovits T, Kovács GM, et al. 2011. Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytologist 191: 789e794. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordeloos ME. 1992. Entoloma s.l. Fungi Europaei, vol. 5. Giovanna Biella, Saronno, Italy. [Google Scholar]
- Noordeloos ME, Morozova OV. 2010. New and noteworthy Entoloma species from the Primorsky Territory, Russian Far East. Mycotaxon 112: 231–255. [Google Scholar]
- Nováková A, Hubka V, Dudová Z, et al. 2014. New species in Aspergillus section Fumigati from reclamation sites in Wyoming (U.S.A.) and revision of A. viridinutans complex. Fungal Diversity 64: 253–274. [Google Scholar]
- O’Donnell K, Cigelnik E, Nirenberg H. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90: 465–493. [Google Scholar]
- O’Donnell K, Lutzoni FM, Ward TJ, et al. 2001. Evolutionary relationships among mucoralean fungi (Zygomycota): Evidence for family polyphyly on a large scale. Mycologia 93: 286–296. [Google Scholar]
- O’Donnell K, McCormick SP, Busman M, et al. 2018. Marasas et al. 1984 ‘Toxigenic Fusarium species: identity and mycotoxicology’ revisited. Mycologia 110: 1058–1080. [DOI] [PubMed] [Google Scholar]
- Ou JH, Lin GC, Chen CY. 2020. Sarocladium species associated with rice in Taiwan. Mycological Progress 19: 67–80. [Google Scholar]
- Peterson SW, Manitchotpisit P, Leathers TD. 2013. Aureobasidium thailandense sp. nov. isolated from leaves and wooden surfaces. International Journal of Systematic and Evolutionary Microbiology 63: 790–795. [DOI] [PubMed] [Google Scholar]
- Pitt JI, Lantz H, Pettersson OV, et al. 2013. Xerochrysium gen. nov. and Bettsia, genera encompassing xerophilic species of Chrysosporium. IMA Fungus 4: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajchenberg M. 1987. Type studies of Polyporaceae (Aphyllophorales) described by J. Rick. Nordic Journal of Botany 7: 553–568. [Google Scholar]
- Rajeshkumar KC, Braun U, Groenewald JZ, et al. 2021. Phylogenetic placement and reassessment of Asperisporium pongamiae as Pedrocrousiella pongamiae gen. et comb. nov. (Mycosphaerellaceae). Fungal Systematics and Evolution 7: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RW. 1970. A Mycological Colour Chart. Commonwealth Mycological Institute, Kew and British Mycological Society. [Google Scholar]
- Réblová M, Untereiner WA, Réblová K. 2013. Novel evolutionary lineagesrevealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS ONE 8: e63547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY. 2000. Towards monophyletic genera in the holomorphic Hypocreales. Studies in Mycology 45: 27–34. [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, et al. 1999. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1–248. [Google Scholar]
- Ryberg M, Nilsson RH, Kristiansson E, et al. 2008. Mining metadata from unidentified ITS sequences in GenBank: a case study in Inocybe (Basidiomycota). BMC Evolutionary Biology 8: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. 1889. Sylloge fungorum omnium hucusque cognitorum VIII. Saccardo, Padova. [Google Scholar]
- Schipper MAA. 1978. On the genera Rhizomucor and Parasitella. Studies in Mycology 17: 52–71. [Google Scholar]
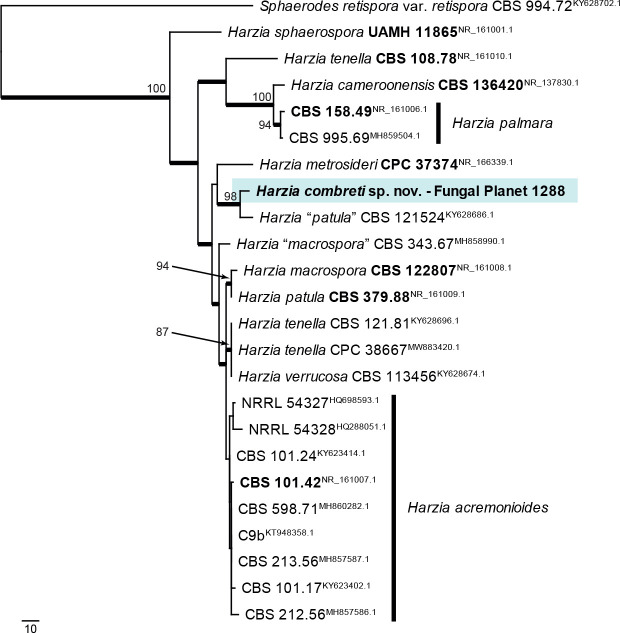
- Schultes NP, Murtishi B, Li DW. 2017. Phylogenetic relationships of Chlamydomyces, Harzia, Olpitrichum, and their sexual allies, Melanospora and Sphaerodes. Fungal Biology 121 (10): 890–904. [DOI] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, et al. 2011. The Genera of Hyphomycetes. CBS Biodiversity Series no. 9: 1–997. CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands. [Google Scholar]
- Shang QJ, Hyde KD, Camporesi E, et al. 2020. Additions to the genus Cytospora with sexual morph in Cytosporaceae. Mycosphere 11: 189–224. [Google Scholar]
- Shearer CA, Crane JL, Chen W. 1999. Freshwater ascomycetes: Ophioceras species. Mycologia 91: 145–156. [Google Scholar]
- Sherwood MA. 1980. Taxonomic studies in the Phacidiales: The genus Coccomyces (Rhytismataceae). Occasional Papers of the Farlow Herbarium of Cryptogamic Botany 15: 1–120. [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical frontend for RAxML. Organisms Diversity and Evolution 12: 335–337. [Google Scholar]
- Singer R. 1947. Contributions towards a monograph of the genus Crepidotus. Lilloa 13: 59–95. [Google Scholar]
- Spotts RA, Seifert KA, Wallis KM, et al. 2009. Description of Cryptosporiopsis kienholzii and species profiles of Neofabraea in major pome fruit growing districts in the Pacific Northwest USA. Mycological Research 113: 1301–1311. [DOI] [PubMed] [Google Scholar]
- Stadler T. 2009. On incomplete sampling under birth-death models and connections to the sampling-based coalescent. Journal of Theoretical Biology 261: 58–66. [DOI] [PubMed] [Google Scholar]
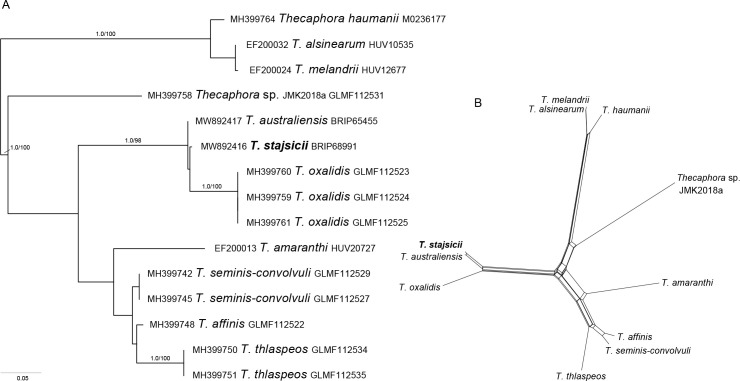
- Stajsic V. 2020a. in VICFLORA: Flora of Victoria, Oxalis: https://vicflora.rbg.vic.gov.au/flora/taxon/0ed554c2-95c9-4cb2-b85e-da75e2655077 [accessed 6 April 2021]. [Google Scholar]
- Stajsic V. 2020b. Oxalidaceae. In: Kodela PG. (ed), Flora of Australia, Australian Biological Resources Study, Canberra. https://profiles.ala.org.au/opus/foa/profile/Oxalidaceae [accessed: 6 April 2021]. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A toll for phylogenetic analysis and post-analysis of large phylogenenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerbell RC, Gueidan C, Guarro J, et al. 2018. The Protean Acremonium. A. sclerotigenum/egyptiacum: Revision, Food Contaminant, and Human Disease. Microorganisms 6: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerbell RC, Gueidan C, Schroers HJ, et al. 2011. Acremonium phylogenetic overview and revision of Gliomastix, Trichothecium and Sarocladium. Studies in Mycology 68: 139–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0b10. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tamura K, Stecher G, Peterson D. et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney JB, Seifert KA. 2020. Mollisiaceae: An overlooked lineage of diverse endophytes. Studies in Mycology 95: 293–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, et al. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research: 44: W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloss RE. 1994. Type studies in Amanita section Vaginatae I: some taxa described in this century (studies 1–23) with notes on description of spores and refractive hyphae in Amanita. Mycotaxon 52: 305–396. [Google Scholar]
- Valenzuela-Lopez N, Cano-Lira JF, Guarro J, et al. 2018. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Studies in Mycology 90: 1–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepol N, Liber J, Desirò A, et al. 2020. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Diversity 104: 267–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vánky K. 2011 ‘2012’. Smut fungi of the world. St. Paul, Minnesota, APS Press. [Google Scholar]
- Vánky K, Lutz M, Bauer R. 2008. About the genus Thecaphora (Glomosporiaceae) and its new synonyms. Mycological Progress 7: 31–39. [Google Scholar]
- Vauras J. 1994. Inocybe diabolica, a new agaric from subarctic regions. Mycologia Helvetica 2: 121–128. [Google Scholar]
- Vauras J, Kokkonen K. 2009. Finnish records of the genus Inocybe. The new species Inocybe saliceticola. Karstenia 48: 57–67. [Google Scholar]
- Vauras J, Larsson E. 2016. Inocybe caprimulgi and I. lacunarum, two new nodulose-spored species from Fennoscandia. Karstenia 55: 1–18. [Google Scholar]
- Vellinga EC. 1988. Glossary. In: Bas C, Kuyper TW, Noordeloos ME, Vellinga EC. (eds). Flora Agaricina Neerlandica vol. 1: 54–64. Blakema, Rotterdam, The Netherlands. [Google Scholar]
- Vellinga EC. 1990. Pluteus. In: Bas C, Kuyper TW, Noordeloos ME, Vellinga EC. (eds), Flora Agaricina Neerlandica, vol 2: 31–55. Blakema, Rotterdam, The Netherlands. [Google Scholar]
- Vellinga EC. 2001. Lepiota. In: Noordeloos ME, Kuyper ThW, Vellinga EC. (eds), Flora Agaricina Neerlandica 5: 109–151. Balkema publishers. [Google Scholar]
- Vellinga EC. 2010. Lepiota in California: species with a hymeniform pileus covering. Mycologia 102: 664–674. [DOI] [PubMed] [Google Scholar]
- Viana FMP, Sousa JA, Araújo JDM, et al. 2012. Fungos Associados a Algumas Espécies Florestais Madeireiras no Distrito de Irrigação de Acaraú-Marco, CE. Documentos / Embrapa Agroindústria Tropical. [Google Scholar]
- Videira SIR, Groenewald JZ, Braun U, et al. 2016. All that glitters is not Ramularia. Studies in Mycology 83: 49–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira SIR, Groenewald JZ, Nakashima C, et al. 2017. Mycosphaerellaceae – Chaos or clarity? Studies in Mycology 87: 257–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Miller OK, Jr. 1983. Biological species in the Collybia dryophila group in North America. Mycologia 75: 707–722. [Google Scholar]
- Visagie CM, Houbraken J. 2020. Updating the taxonomy of Aspergillus in South Africa. Studies in Mycology 95: 253–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli M, Vessella F, Cardoni S, et al. 2017. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). Tree Genetics & Genomes 13.3. [Google Scholar]
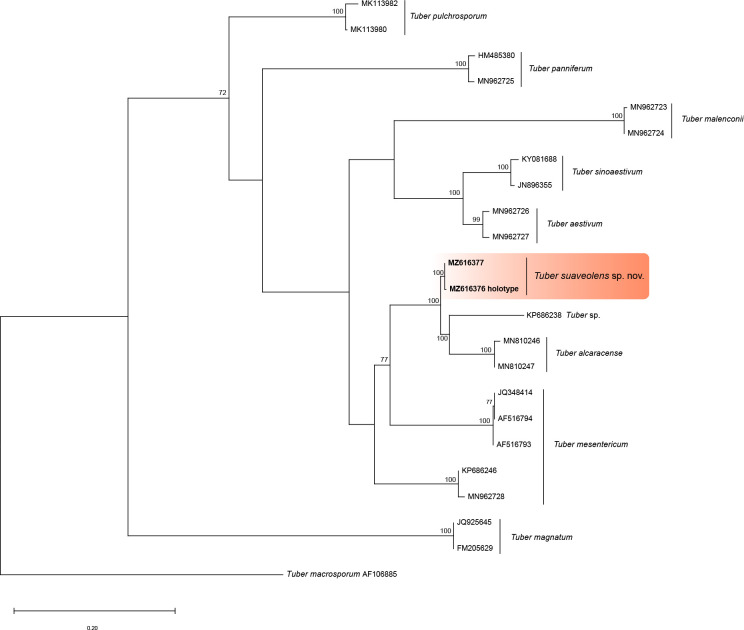
- Vittadini C. 1831. Monographia Tuberacearum. Typographia Rusconi, Milano, Italy. [Google Scholar]
- Vizzini A, Angelini C, Ercole E. 2017. Is the species diversity in the lyophylloid genera Calocybella and Gerhardtia (Agaricales, Basidiomycota) underestimated? Two new species from the Dominican Republic. Phytotaxa 291: 241–252. [Google Scholar]
- Vizzini A, Consiglio G, Setti L, et al. 2015. Calocybella, a new genus for Rugosomyces pudicus (Agaricales, Lyophyllaceae) and emendation of the genus Gerhardtia. IMA Fungus 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzini A, Liang JF, Jančovičová S, et al. 2014. Lepiota coloratipes, a new species for Lepiota rufipes ss. Auct. europ. non ss. orig. Mycological Progress 13: 171–179. [Google Scholar]
- Vizzini A, Tatti A, Huijser HA, et al. 2019. Looking for Lepiota psalion Huijser & Vellinga (Agaricales, Agaricaceae). MycoKeys 52: 45–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzini A, Zotti M, Traverso M, et al. 2016. Variability, host range, delimitation and neotypification of Amanita simulans (Amanita section Vaginatae): collections associated with Helianthemum grasslands, and epitypification of A. lividopallescens. Phytotaxa 280: 1–22. [Google Scholar]
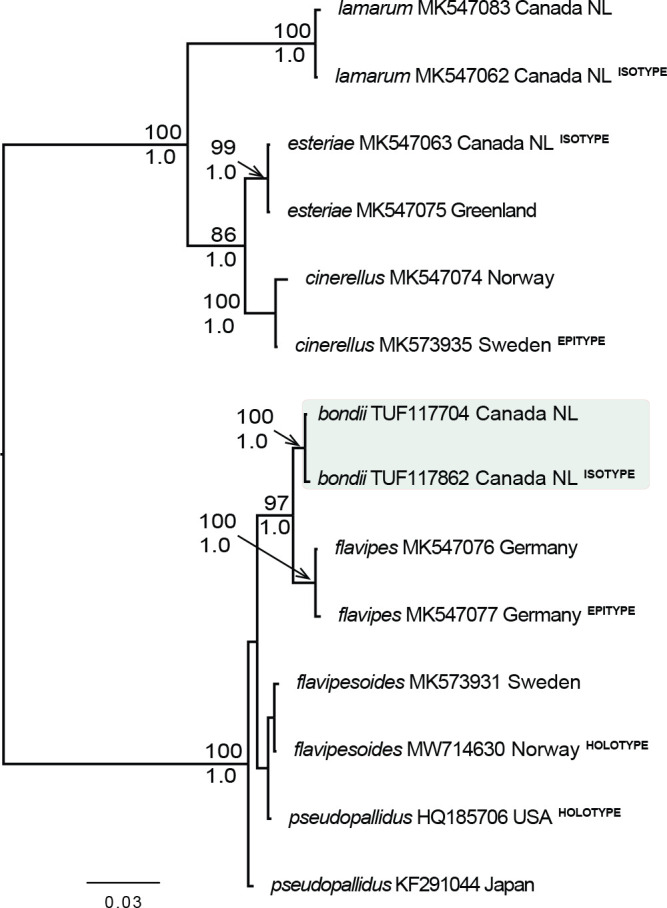
- Voitk A, Saar I, Lodge DJ, et al. 2020a. From Esteri’s to Bond’s mushroom — some new and old Cuphophylli in NL. Omphalina 11: 71–80. [Google Scholar]
- Voitk A, Saar I, Lodge DJ, et al. 2020b. New species and reports of Cuphophyllus from northern North America compared with related Eurasian species. Mycologia 112: 438–452. [DOI] [PubMed] [Google Scholar]
- Vu D, Groenewald M, Szöke S. et al. 2016. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Studies in Mycology 85: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Jones EBG, Camporesi E, et al. 2015. The genus Murispora. Cryptogamie Mycologie 36: 419–448. [Google Scholar]
- Wang H, Gu ZR, Zhao CL. 2021. Hyphodermella zixishanensis (Polyporales, Basidiomycota), a new species with reddish hymenial surface. Nordic Journal of Botany 39: e03329. [Google Scholar]
- Wu WP, Zhuang WY. 2005. Sporidesmium, Endophragmiella and related genera from China. Fungal Diversity Research Series 15: 1–351. [Google Scholar]
- Yilmaz N, Sandoval-Denis M, Lombard L, et al. 2021. Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 46: 129–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H-S, Dai Y-C. 2012. Wood-inhabiting fungi in southern China 5. Polypores from Guangxi Autonomous Region. Annales Botanici Fennici 49: 341–351. [Google Scholar]
- Yurkov A, Guerreiro MA, Sharma L. et al. 2015. Multigene assessment of the species boundaries and sexual status of the basidiomycetous yeasts Cryptococcus flavescens and C. terrestris (Tremellales). PLoS One 10: e0120400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalar P, Gostinčar C, De Hoog GS, et al. 2008. Redefinition of Aureobasidium pullulans and its varieties. Studies in Mycology 61: 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller SM, Smith AH. 1964. The genus Calvatia in North America. Lloydia 27: 148–186. [Google Scholar]
- Zhao R-L, Desjardin DE, Soytong K, et al. 2010. A monograph of Micropsalliota in northern Thailand based on morphological and molecular data. Fungal Diversity 45: 33–79. [Google Scholar]

































































 i, Vietnamese vernacular name for Musa spp., from which the ex-type strain was isolated.
i, Vietnamese vernacular name for Musa spp., from which the ex-type strain was isolated. An Province, Con Cuông District, Châu Khê commune, N19°1’48.73" E104°43’31.97", inside seed of M. itinerans, 18 Nov. 2014, L.T. Phong, V.V. Tung & T.T. Duong, isol. R. Hill (culture CBS 148465; ITS, LSU, cmdA, rpb1, rpb2, tef1 and tub2 sequences GenBank
An Province, Con Cuông District, Châu Khê commune, N19°1’48.73" E104°43’31.97", inside seed of M. itinerans, 18 Nov. 2014, L.T. Phong, V.V. Tung & T.T. Duong, isol. R. Hill (culture CBS 148465; ITS, LSU, cmdA, rpb1, rpb2, tef1 and tub2 sequences GenBank 















 δωμα-, railing, and - ϕ
δωμα-, railing, and - ϕ