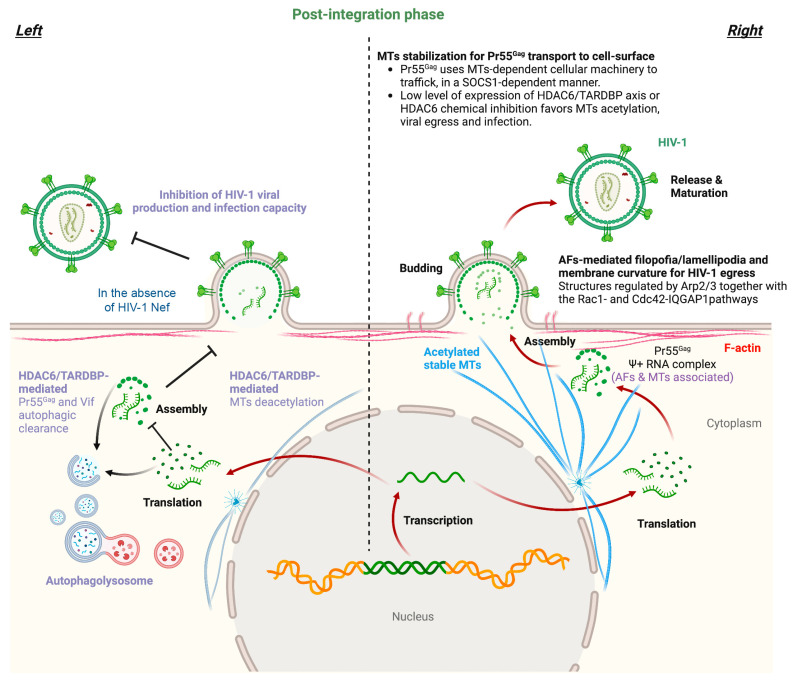
Figure 4.
Scheme illustrating the late stages of the HIV-1 life cycle and associated tubulin and actin cytoskeleton dynamics. The transcription and translation of the integrated HIV-1 genome generate viral RNA+ and proteins, with the structural Pr55Gag polyprotein being relevant to recognize viral RNA+ and recruit a complex to stable MTs to travel to the plasma membrane (right panel). In fact, HIV-1 Pr55Gag uses MTs-dependent cellular machinery to traffic to the cell surface in a SOCS1-dependent manner. Pr55Gag also interacts with F-actin, thus assembling at the cell surface in areas where AFs are remodeled by the Arp2/3- and Rac1/Cdc42/IQGAP1 pathways, inducing F-actin filopodia and lamellipodia structures that finally generate high positive membrane curvatures where HIV-1 egresses (right panel). The MT-associated HDAC6 enzyme deacetylates stable MTs, thereby impeding Pr55Gag cell surface localization (left panel). Likewise, HDAC6 tubulin-deacetylase function is required to target HIV-1 Pr55Gag and Vif proteins for autophagy degradation, thus inhibiting viral production and infection capacity (left panel). HIV-1 Nef counteracts these antiviral actions by targeting the MT-associated enzyme, thereby stabilizing MTs and the HIV-1 Pr55Gag and Vif proteins. Therefore, HIV-1 Nef promotes the transport of Pr55Gag to the cell surface where it assembles to form viral particles that incorporate Vif to ensure the infectivity of HIV-1. Designs and templates were created with BioRender.