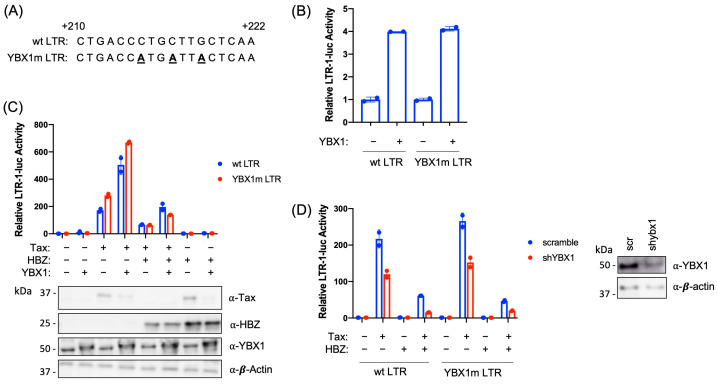
Figure 6.
Mutation of the proposed YBX1 binding site does not affect viral transcription. (A) The proposed YBX1 binding site was mutated in the LTR-1-luciferase expression vector through site-directed mutagenesis (shown as bold, underlined text). An alignment of the YBX1 mutation (YBX1m) region and the wt region from the HTLV-1 LTR is shown. (B) HEK293T cells were co-transfected with pcDNA3.1(+) empty, YBX1, wt HTLV-1 LTR-firefly luciferase, or YBX1m HTLV-1 LTR-firefly luciferase, and tk-renilla luciferase (internal control) constructs, as indicated. At 48 h post-transfection, cells were collected for luciferase assays to measure relative LTR transactivation. The empty vector was set at 1. (C) HEK293T cells were co-transfected with pcDNA3.1(+) empty, YBX1, HBZ, Tax, HTLV-1 LTR-firefly luciferase, YBX1m LTR-firefly luciferase, and tk-renilla luciferase (internal control) constructs, as indicated. At 48 h post-transfection, cells were collected for luciferase assays to measure relative LTR transactivation (top panel). The empty vector was set at 1. Total cell lysates were examined by immunoblot analysis using antibodies to YBX1, HBZ, Tax, and β-actin (bottom panel). (D) HEK293T scramble and shYBX1 cells were co-transfected with pcDNA3.1(+) empty, Tax, HBZ, HTLV-1 LTR-firefly luciferase, YBX1m LTR-firefly luciferase, and tk-renilla luciferase (internal control) constructs, as indicated. At 48 h post-transfection, cells were collected for luciferase assays to measure relative LTR transactivation. The empty vector was set at 1. Total cell lysates were examined by immunoblot analysis using antibodies to YBX1 and β-actin (right panel). Graphs represent data generated from duplicate samples, and error bars represent the standard deviation (SD). The data are representative of at least three experimental repeats.