Figure 7.
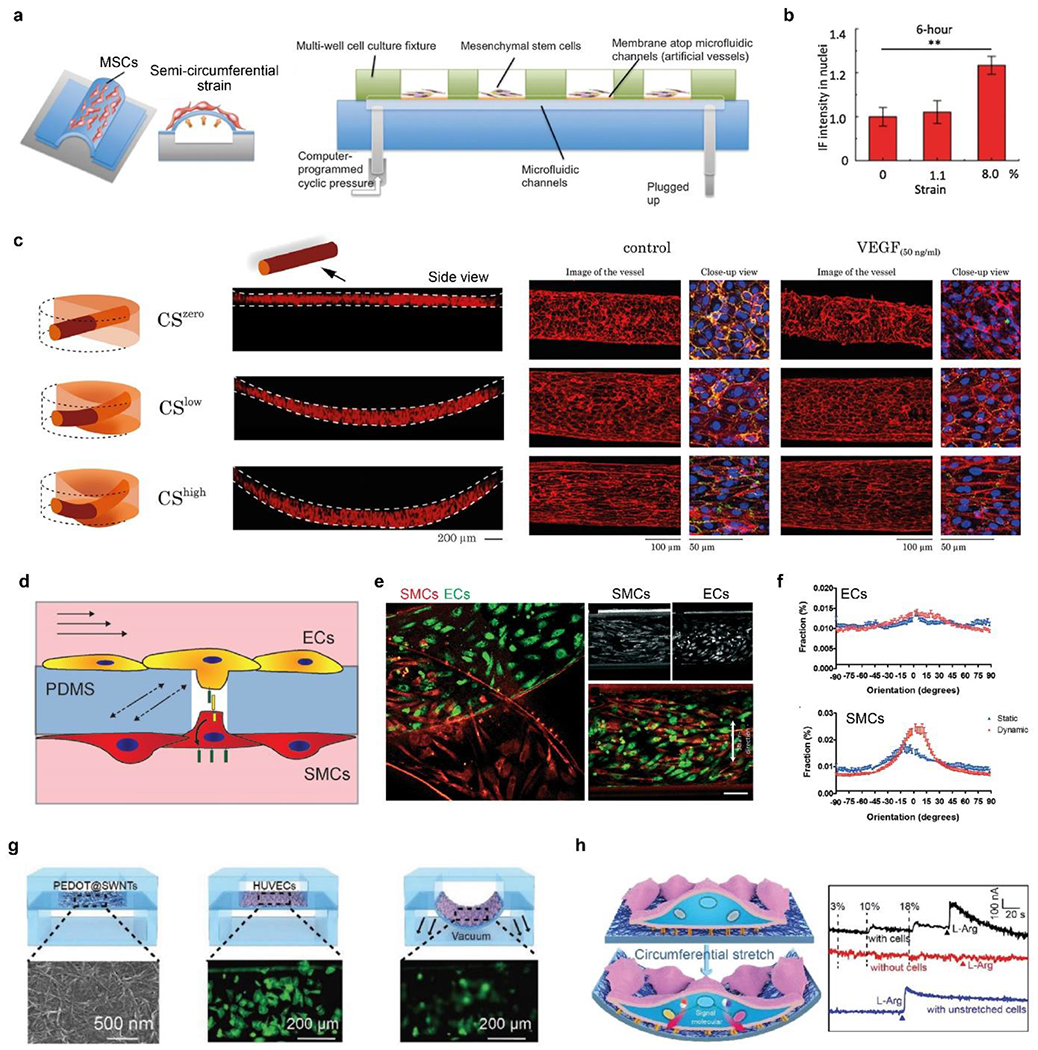
a) Schematic of microfluidic chips with semi-circumferential deformation. b) quantitative comparison of the nucleus accumulation of β-catenin under circumferential strains. a-b) Reproduced with permission.[254] Copyright 2012, Oxford University Press. c) Schematic and fluorescence images (side view) of the 3D vessels under zero, low, and high circumferential strains (CS). CS increases cellular alignment and counteracts VEGF to stabilize vessels. F-actin (red), platelet endothelial cell adhesion molecule-1 (PECAM-1, green), and DAPI (blue). Reproduced with permission.[257] Copyright 2021, Creative Commons Attribution license. d) Schematic of ECs and SMCs cocultured on a porous PDMS membrane in a vessel chip and exposed to shear stress and circumferential strain. e) Fluorescence image of ECs and SMCs after 4-day of culturing under mechanical stimuli, i.e., dynamic culture. Scale bar, 100 μm. f) Quantitative analyses of cellular alignment under static and dynamic culturing conditions. d-f) Reproduced with permission.[258] Copyright 2018, Creative Commons Attribution license. g) Schematic of in situ, real-time monitoring of mechanical stimuli using a flexible electrochemical sensor. HUVECs (in green) become blurred when they stretched and were out of focus. H) Right, schematic of cellular mechanotransduction to release NO. Left, electrochemical curves of cells responding to 0%, 10%, and 18% strains, and L-arginine. G-h) Reproduced with permission.[262] Copyright 2019, John Wiley and Sons.
