Figure 1.
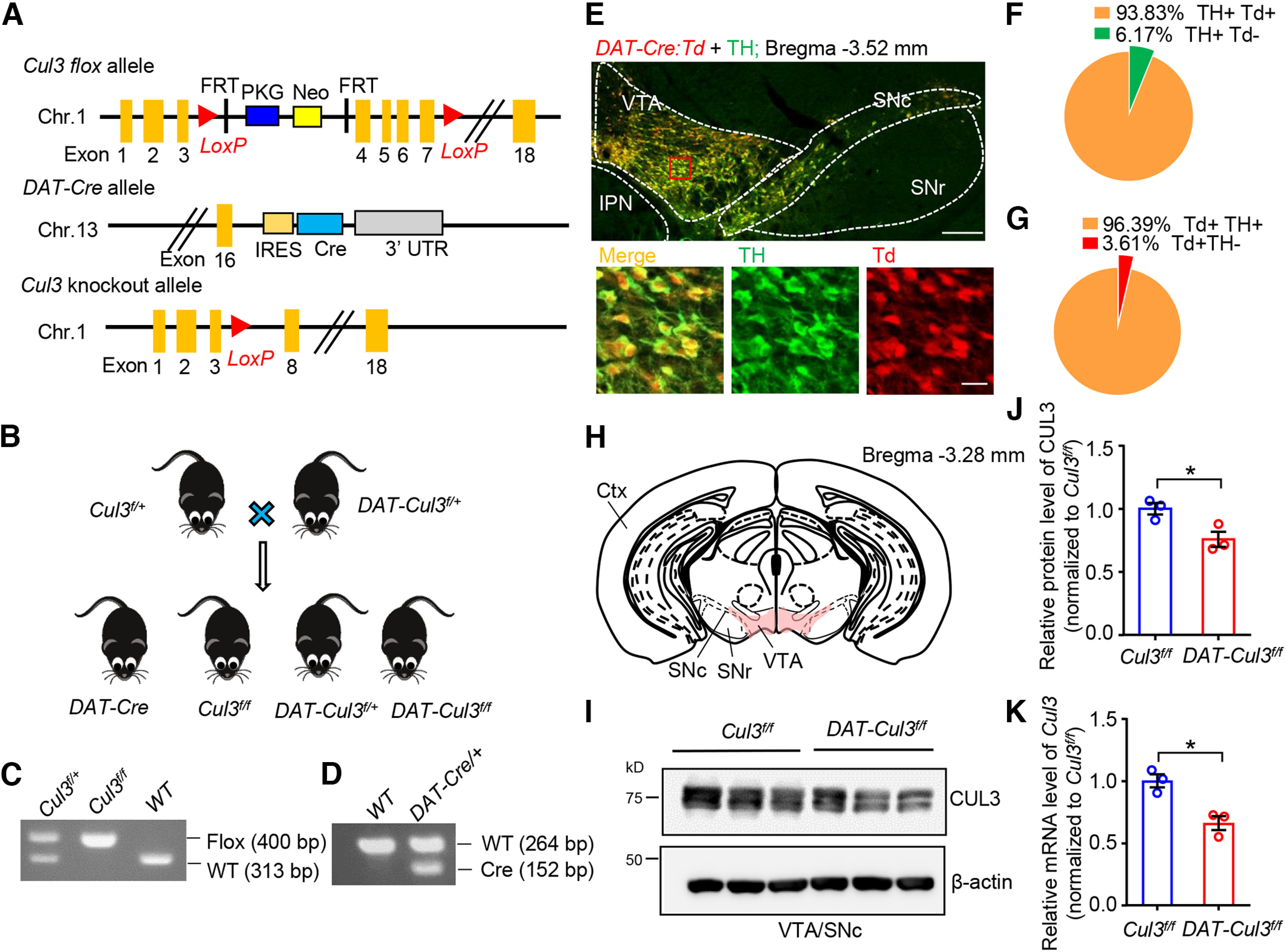
Generation and characterization of DAT-Cul3 conditional KO mice. A, Schematic diagram of the genomic structure of Cul3 flox, DAT-Cre, and Cul3 KO alleles in mice. Chr., chromosome. FRT, flippase recognition target. IRES, internal ribosome entry site. LoxP, locus of X-over of P1. Neo, neomycin phosphotransferase gene. PKG, phosphoglycerate kinase I promoter. UTR, untranslated region. B, Genetic strategy for ablation of Cul3 in DA neurons. Cul3f/+ mice were crossed with DAT::Cre::Cul3f/+ (referred to as DAT-Cul3f/+) to get DAT-Cul3f/f mice together with other indicated genotypes. C, D, Representative PCR genotyping results for Cul3 allele (C) and DAT-Cre allele (D). E, Representative images of mice midbrain from DAT::Cre::Ai9 (DAT-Cre:Td) mouse at P60. The coronal section of the midbrain was stained with TH (visualized by AlexaFluor-488, green). tdTomato (Td) signals were imaged without further staining. Scale bar, 200 μm. Bottom, Higher magnification of the inset. Scale bar, 20 μm. The Td signals driven by DAT-Cre were restricted to TH-positive DA neurons. IPN, interpeduncular nucleus. F, Ratio of Td/TH double-positive (Td+ TH+) in TH+ neurons in VTA/SNc. G, Ratio of Td/TH double-positive (Td+ TH+) in Td+ neurons in VTA/SNc. H, Schematic diagram of dissecting VTA and part of SNc (VTA/SNc, in pink) on coronal sections for Western blot and real-time PCR assays. Ctx, Cortex; SNr, substantia nigra pars reticulata. I, Reduced CUL3 protein in VTA/SNc in CUL3-deficient mice at P60. Homogenates of VTA/SNc were subjected to Western blotting. J, Quantification for data in I. n = 3 mice per group; Cul3f/f versus DAT-Cul3f/f, p = 0.0375, t(3.709) = 3.175; unpaied t test. K, Reduced mRNA levels of Cul3 determined by real-time PCR for VTA/SNc in DAT-Cul3f/f mice compared with Cul3f/f controls at P60. n = 3 mice per group; Cul3f/f versus DAT-Cul3f/f, p = 0.0118, t(3.981) = 4.403; unpaied t test. Data are mean ± SEM. *p < 0.05.
