Figure 3.
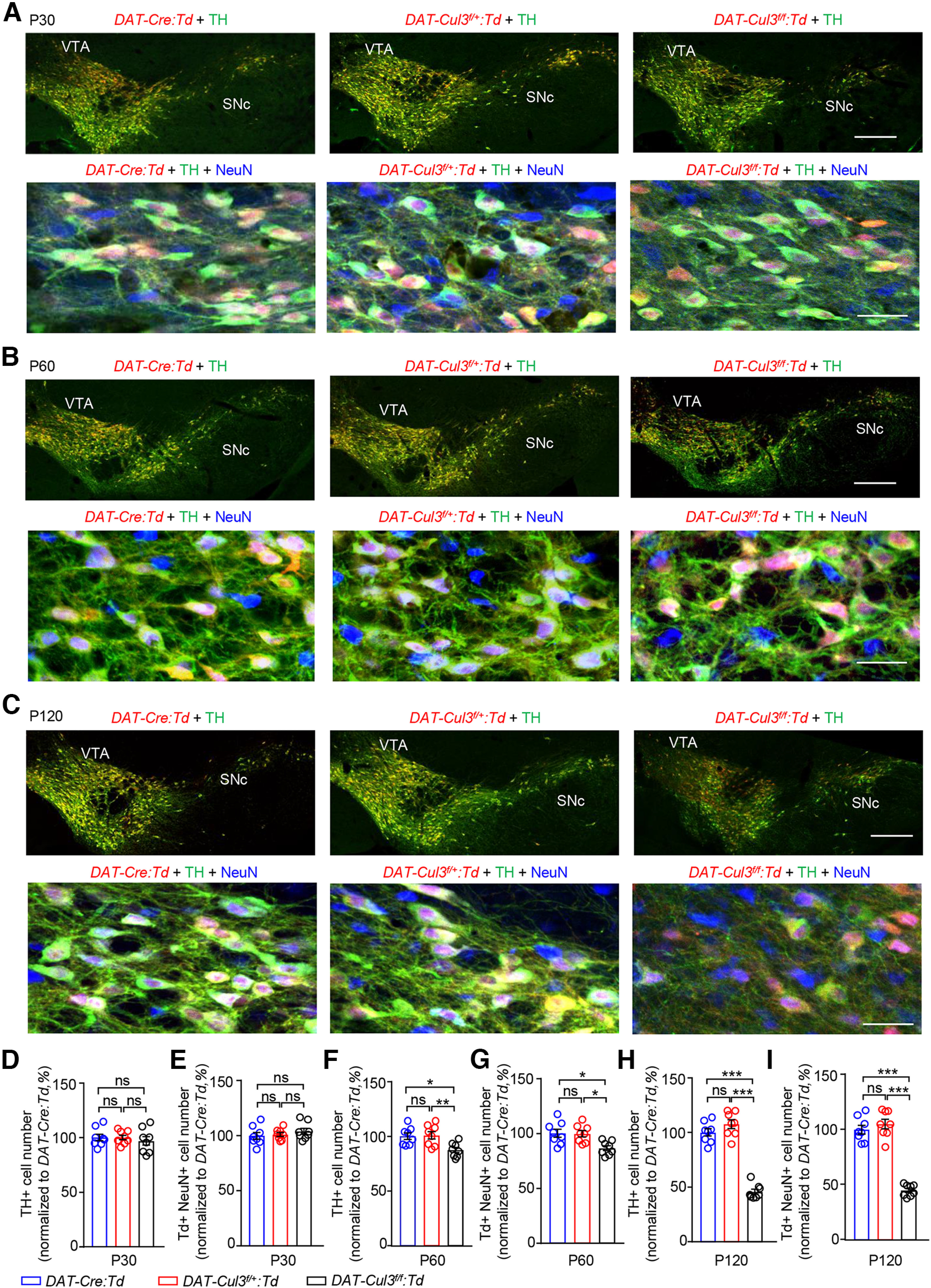
Comparable numbers of midbrain DA neurons in adult DAT-Cul3f/+ mice. A, Similar number of midbrain DA neurons in P30 DAT-Cul3f/+:Td and DAT-Cul3f/f:Td mice compared with controls (DAT-Cre:Td). Top, Representative images for VTA/SNc in coronal sections of the midbrain at bregma −3.5 mm. Sections were stained with TH antibodies (green) and imaged directly for the Td (red) signal. Scale bar, 200 μm. Bottom, Enlarged images of VTA for indicated genotypes. Coronal sections were stained with TH (green) and NeuN (blue) antibodies, and Td (red) signals were imaged without further staining. Scale bar, 20 μm. B, Decreased VTA/SNc DA neurons in P60 DAT-Cul3f/f:Td mice, but not DAT-Cul3f/+:Td mice, compared with controls (DAT-Cre:Td). Top, Representative images for VTA/SNc in coronal sections of the midbrain at bregma −3.53 mm. Sections were stained with TH antibodies (green) and imaged directly for the tdTomato (Td, red) signal. Scale bar, 200 μm. Bottom, Enlarged images of VTA for indicated genotypes. Coronal sections were stained with TH (green) and NeuN (blue) antibodies, and Td (red) signals were imaged without further staining. Scale bar, 20 μm. C, Decreased VTA/SNc DA neurons in P120 DAT-Cul3f/f:Td mice, but not DAT-Cul3f/+:Td mice, compared with controls (DAT-Cre:Td). Top, Representative images for VTA/SNc in coronal sections of the midbrain at bregma −3.5 mm. Sections were stained with TH antibodies (green) and imaged directly for the tdTomato (Td, red) signal. Scale bar, 200 μm. Bottom, Enlarged images of VTA for indicated genotypes. Coronal sections were stained with TH (green) and NeuN (blue) antibodies, and Td (red) signals were imaged without further staining. Scale bar, 20 μm. D, E, Similar number of TH+ (D) and Td+ NeuN+ cells (E) in VTA/SNc of DAT-Cul3f/f:Td and DAT-Cul3f/+:Td mice compared with controls (DAT-Cre:Td) at P30. n = 8 slices from 4 mice per genotype. E, DAT-Cul3f/+:Td versus DAT-Cre:Td, p > 0.9999; DAT-Cul3f/f: Td versus DAT-Cre:Td, p = 0.7605; DAT-Cul3f/f: Td versus DAT-Cul3f/+:Td, p = 0.7670; F(2,21) = 0.3306. F, DAT-Cul3f/+:Td versus DAT-Cre:Td, p = 0.9455; DAT-Cul3f/f: Td versus DAT-Cre:Td, p = 0.5173; DAT-Cul3f/f: Td versus DAT-Cul3f/+:Td, p = 0.7113; F(2,21) = 0.6559. F, G, Reduced number of TH+ (F) and Td+ NeuN+ cells (G) in VTA/SNc of DAT-Cul3f/f:Td mice, but not DAT-Cul3f/+:Td mice compared with control (DAT-Cre:Td) at P60. n = 8 slices from 4 mice per genotype. G, DAT-Cul3f/+:Td versus DAT-Cre:Td, p = 0.9785; DAT-Cul3f/f: Td versus DAT-Cre:Td, p = 0.0149; DAT-Cul3f/f: Td versus DAT-Cul3f/+:Td, p = 0.0095; F(2,21) = 6.78. H, DAT-Cul3f/+:Td versus DAT-Cre:Td, p = 0.9916; DAT-Cul3f/f: Td versus DAT-Cre:Td, p = 0.0102; DAT-Cul3f/f: Td versus DAT-Cul3f/+:Td, p = 0.0135; F(2,21) = 6.788. H, I, Reduced number of TH+ (H) and Td+ NeuN+ cells (I) in VTA/SNc of DAT-Cul3f/f: Td mice, but not DAT-Cul3f/+:Td mice compared with control (DAT-Cre:Td) at P120. n = 8 brain slices from 4 mice per genotype. I, DAT-Cul3f/+:Td versus DAT-Cre:Td, p = 0.2569; DAT-Cul3f/f: Td versus DAT-Cre:Td, p < 0.0001; DAT-Cul3f/f: Td versus DAT-Cul3f/+:Td, p < 0.0001; F(2,21) = 113.2. J, DAT-Cul3f/+:Td versus DAT-Cre:Td, p = 0.5695; DAT-Cul3f/f: Td versus DAT-Cre:Td, p < 0.0001; DAT-Cul3f/f: Td versus DAT-Cul3f/+:Td, p < 0.0001; F(2,21) = 98.19; one-way ANOVA followed by Tukey's post hoc test. Data are mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001.
